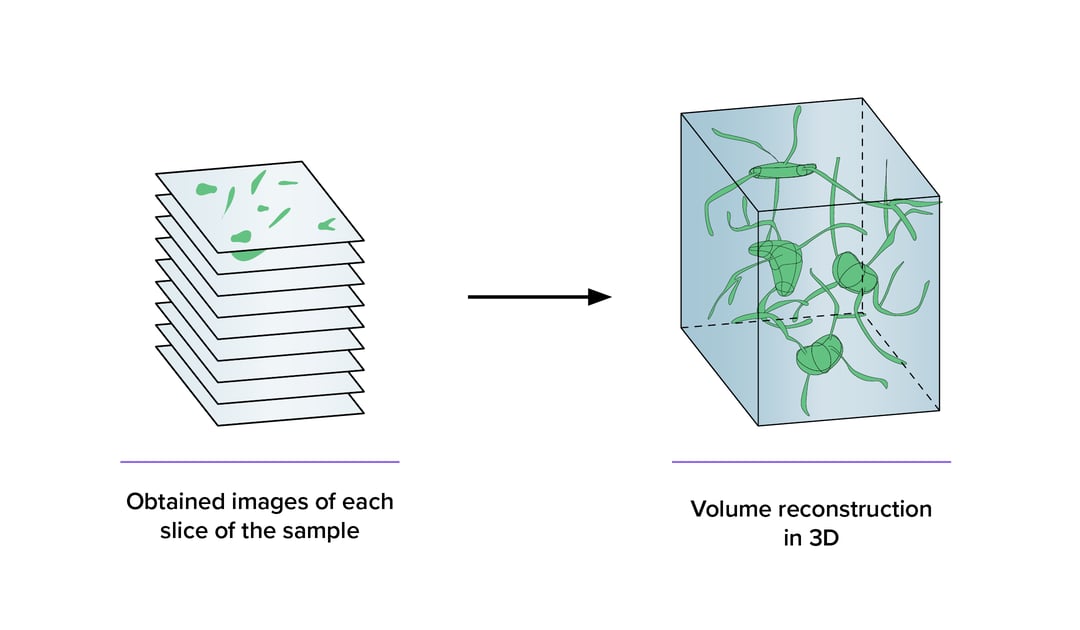
However, most of the existing imaging technologies typically produce a 2D images, which are challenging to use for 3D data reconstruction.
Nowadays, a big variety of 3D imaging techniques are available. One such is light microscopy that obtains 3D images of cell structures and provides valuable information on structure-function relationships in cells and tissues. The drawback of light microscopy techniques is the need for chemical markers that are limited since they are target specific, and the development of markers is hard and tedious. To overcome these drawbacks, scanning electron microscopy (SEM) can be used instead [1].
Nevertheless, without modifications a traditional SEM does not capture 3D information. Furthermore, samples serial sectioning with hundreds of thin slices tremendously increased sample handling time. These drawbacks lead to the software and hardware improvements to capture 3D information in an automated mode. As a result, these automated SEM approaches emerged: serial block-face (SBEM), focused ion beam SEM (FIB-SEM), and array tomography [2]. The difference between these SEM techniques is the means with which serial sectioning is achieved. SBEM uses an ultramicrotome integrated in the vacuum chamber, that automatically removes a section from the specimen after scanning the surface with the electron beam. A similar approach is implemented in FIB-SEM, but instead of using a diamond knife to expose the new surface, a focused ion beam is used. Finally, in array tomography, sections from the sample are collected on a conductive substrate like a wafer, which is then imaged in a SEM. This has the benefit of leaving samples intact for reimaging at a later point, while SBF-SEM and FIB-SEM destroy the sample during the imaging process.
All the mentioned SEM techniques are challenging and require a lot of experience. Most of all, it is time-consuming to obtain data of a large volume specimens with a single electron beam. For instance, in neurobiology, collecting the EM data required to map all neuronal connections in a small creature like a mouse would already take years [3]. Therefore, there is a high demand for faster SEM methods. One of the solutions is an SEM capable of imaging with multiple electron beams simultaneously. At Delmic, we are currently developing such a multi-beam solution, where scanning EM is combined with an optical transmission detector (optical STEM). This STEM approach allows fast and reliable imaging of the large volume biological specimen with nanometer resolution.
Volume microscopy has a wide range of applications as there is a need to understand the ultrastructural details of biological processes and functions. For example, in plant physiology, it is of high importance to assign molecular roles to the corresponding gene and determine their overall contributions in plant processes and structure [4]. Therefore, obtained data on structural differences is analysed together with available genome data to decipher genetic or functional data within an ultrastructural context. The volume microscopy also found application in histology-pathology for diagnostic purposes studies, where tissue architecture is examined in bulk [5].
In conclusion, electron volume microscopy is a valuable tool that helps to reveal structure-function relationships of the complex biological systems. With Delmic’s current development of fast electron microscopy, we aim to provide a stable, automated workflow based around minimal operator intervention. Moreover, Delmic aims to make volume microscopy techniques available to more research areas such as biofilm analysis, food engineering, and diagnostics.
To learn more about the possibilities of fast electron microscopy and Delmic's solution, please visit our fast EM fundamentals section.
References
[1] C. J. Peddie and L. M. Collinson, “Exploring the third dimension: Volume electron microscopy comes of age,” Micron, vol. 61, pp. 9–19, 2014, doi: 10.1016/j.micron.2014.01.009.
[2] B. Titze and C. Genoud, “Volume scanning electron microscopy for imaging biological ultrastructure,” Biol. Cell, vol. 108, no. 11, pp. 307–323, 2016, doi: 10.1111/boc.201600024.
[3] K. L. Briggman and D. D. Bock, “Volume electron microscopy for neuronal circuit reconstruction,” Curr. Opin. Neurobiol., vol. 22, no. 1, pp. 154–161, 2012, doi: 10.1016/j.conb.2011.10.022.
[4] W. Shen et al., “Three-dimensional reconstruction of Picea wilsonii Mast. pollen grains using automated electron microscopy,” Sci. China Life Sci., vol. 63, no. 2, pp. 171–179, 2020, doi: 10.1007/s11427-019-9820-4.
[5] N. Roberts et al., “Toward Routine Use of 3D Histopathology as a Research Tool,” Biophys. Imaging Comput. Biol., vol. 180, no. 5, pp. 1835–1842, 2012, doi: 10.1016/j.ajpath.2012.01.033.
.png)