Electron microscopy is being widely used to examine the structures of cells, tissues and organs, and to bring the researchers closer to answering fundamental questions. Thanks to its high resolution, even small morphological differences can be identified and compared.
This strength has been used in several studies to provide novel insights in a variety of diseases. Typically, cell lines or tissues from different donors are screened by EM, after which the morphology of experimental conditions is compared with the control specimen. For example, analysing whole pancreatic islets from donors with type 1 diabetes, and comparing the results with healthy tissues, can bring us closer to understanding the pathophysiological mechanisms of the disease [1]. Therefore, quantitatively comparable large-scale EM data can be extremely beneficial for understanding the nature of diseases such as diabetes, pemphigus [2] or eosinophilic esophagitis [3], and many more. Large-scale EM also finds application in studying consequences and origins of cancer cells. Important conclusions can be made by systematically comparing cells from different stages of tumour progression, which can contribute to the development of clinical applications [4].
However, the approach is not yet without drawbacks
There are several limitations to the projects that aim to acquire both macromolecular details and tissue context, and even more when multiple samples have to be compared.
Conventional screening of samples leads to several challenges. First is of all, it is extremely labor-intensive, as sample screening and image acquisition are done manually. If specific features across different samples have to be compared, the process has to be repeated multiple times. Features of interest might not be visible in a section at all, since a section represents only a tiny volume of a cell or tissue. This means that acquiring statistically relevant data through a traditional, manual EM workflow alone can be an extremely time-consuming or even unfeasible prospect.
Another important downside of the conventional manual approach comes as a result of the previous one. Often the research has to be limited to capture only the structures of interest at a high resolution, while acquiring the rest of the data at a low magnification. This leads to having limited context, which in its turn limits holistic evaluation of the sample. Finally, if more images from other regions of the sample are needed, the operator has to reacquire images and spend even more time at the microscope.
Fast electron microscopy brings much needed automation and high throughput to the table
It’s the ability to get both complete overview and in-depth data at a macromolecular resolution of a tissue that makes fast electron microscopy so attractive. A high-speed electron microscope combined with an automated workflow can eliminate the bottleneck created by the throughput of conventional EMs. Sections can be imaged in their entirety, after which the full, high-resolution dataset is available for (automated) analysis. In addition, this way data is acquired in an unbiased manner: datasets contain the full context of the cell or tissue, rather than a context provided by a (possibly) biased researcher.
Delmic has been working on a high-throughput system which is capable of automated data acquisition for quantitative analysis, from a wide range of samples. Being able to deliver powerful insights while keeping the workflows simple, this system allows to shift the focus from operating the microscope to analysing the data.
Would you like to learn how this solution can help your research? Then watch our webinar to learn more and participate in the discussion.
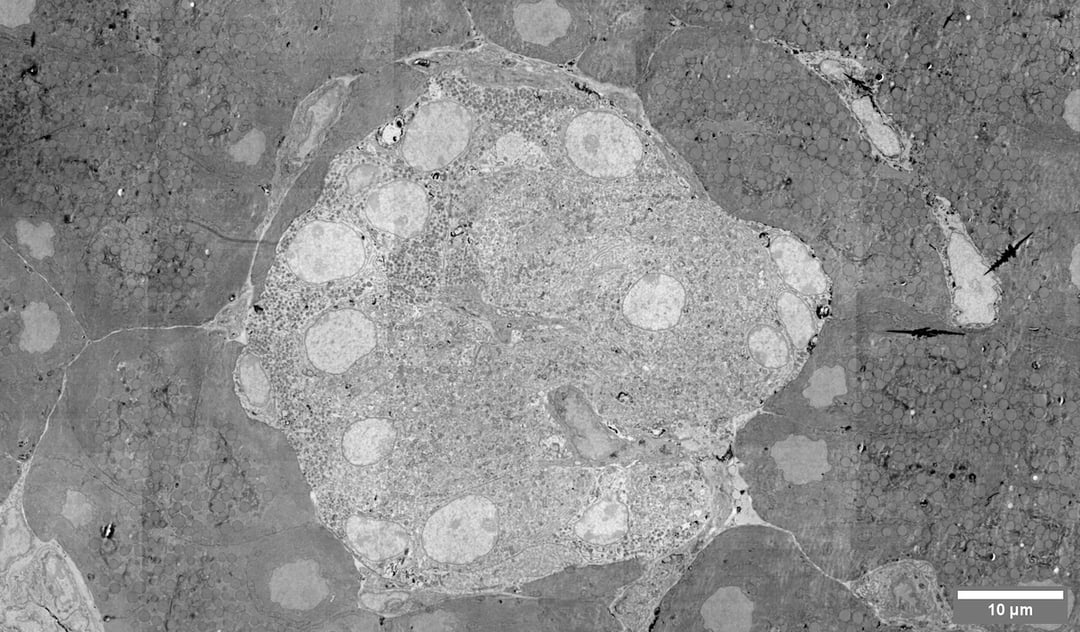
Image: megafield of a pancreatic islet, captured on Delmic's Fast EM prototype. Morphological analysis of islets can help us understand the pathophysiological mechanisms of diabetes. Sample courtesy of Ben Giepmans' group, Department of Cell Biology, University Medical Center Groningen.
References:
[1] de Boer, P., Pirozzi, N.M., Wolters, A.H.G. et al. (2020). Large-scale electron microscopy database for human type 1 diabetes. Nat Commun 11, 2475. https://doi.org/10.1038/s41467-020-16287-5
[2] Sokol, E., Kramer, D., Diercks, G. F. H., Kuipers, J., Jonkman, M. F., Pas, H. H., & Giepmans, B. N. G. (2015). Large-scale electron microscopy maps of patient skin and mucosa provide insight into pathogenesis of blistering diseases. Journal of Investigative Dermatology, 135(7), 1763–1770. https://doi.org/10.1038/jid.2015.109
[3] Saffari, H., Hoffman, L. H., Peterson, K. A., Fang, J. C., Leiferman, K. M., Pease III, L. F., Gleich, G. J. (2014). Electron microscopy elucidates eosinophil degranulation patterns in patients with eosinophilic esophagitis. The Journal of Allergy and Clinical Immunology, 133(6), 1728-1734. https://doi.org/10.1016/j.jaci.2013.11.024
[4] Marteil, G., Guerrero, A., Vieira, A.F. et al. (2018). Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat Commun 9, 1258. https://doi.org/10.1038/s41467-018-03641-x
.png)