With limited imaging resources, providing every project stakeholder with the data needed to answer their research question becomes a heavy task and can lead to a backlog of projects. Facility operators may feel pressured, and projects may be turned down due to a lack of available resources or time. The situation is further complicated by a growing need in the research community to image larger and more intricate samples, such as whole organs or bulk tissue. So, what types of projects require a lot of time in imaging facilities, and how does a fast imaging solution help them out?
How can high-throughput imaging benefit imaging facilities?
In a facility setting high-throughput electron imaging is beneficial for multiple reasons. It gets results to stakeholders faster, can be used to image more or larger samples, and helps users to quantitatively compare data of multiple specimens. But how do we achieve higher throughput in electron imaging, exactly?
One part of this, of course, is improving the speed with which an image is collected by using a faster detector or a camera, for example. Another aspect is making sure a system can rapidly process many samples by efficient sample exchange or the ability to load multiple samples. With a sufficiently fast system samples can be imaged rapidly in their entirety, after which data analysis is done offline. Operators, therefore, no longer need to spend substantial amounts of time behind the microscope to search for the structure of interest, leaving the system available for data collection, and the operator free to perform analysis or support their facility. Overall, this approach can drastically reduce the time between handing in samples and getting back results, which can be crucial for facilities that also service pathology or diagnostics labs.
An added benefit of this approach is that it serves as a bias-free method of recording data: samples are imaged in their entirety, rather than specific parts relevant only to the research question. This is particularly useful for comparative studies, where multiple samples are required to be imaged and compared to observe healthy and diseased conditions, or the effects of drug treatments or genetic modification. This can provide new unique insights into diseases [1] and do so significantly faster.
Volume electron microscopy
For many projects within an imaging facility two-dimensional data is enough to answer a research question. However, processes in life rarely occur in two dimensions, so 3D imaging is needed to fully understand the interactions in a tissue or an organism. Several research fields therefore heavily rely on volume electron microscopy (EM) techniques to examine the ultrastructure of their sample. In a general sense, volume EM projects share a common challenge: a requirement for nanoscale detail in a large-scale context. Projects like these may require weeks or months of continuous imaging on a conventional EM, leaving it unavailable for other research projects. Some examples are detailed below.
In cell biology and structural biology, mapping of cells, tissues and organs through volume EM provides insight in tissue architecture and the interactions taking place between different cell types. These ultrastructural datasets are often used as a data point on their own, or serve as the basis for more complex imaging approaches. For example, an ultrastructural map of a whole organism provides context for observations made in fluorescence microscopy and helps understand the patterns of gene expression seen in model organisms [2, 3]. However, even small model animals like C. elegans or Platynereis dumerilii require hundreds of thousands of image tiles to image completely [3].
In neurobiology, ultrastructural data is often used for connectomics, a research field aiming to map neural connections to understand behaviour, learning and disease. EM has become a workhorse in connectomics, as it is the only technique suitable for unambiguously identifying neurons and synapses at nanometer resolution. However, imaging large volumes at such high resolution creates a unique challenge in terms of data collection. Even a drosophila brain (with a volume of <1 mm3) already generates hundreds of terabytes worth of data [4]. For larger model systems, both the time required, and the data produced may be manifold bigger. Speeding up the acquisition of data through high-throughput imaging systems is therefore highly beneficial within an imaging facility.
Delmic is currently working on a powerful fast electron microscopy solution which, thanks to its high sustained throughput, can significantly improve the output of imaging facilities, and the quantity and quality of data that can be acquired to answer research questions. This solution makes projects possible that were previously unfeasible due to the volume of an individual specimen, or because the project requires comparison of many individuals.
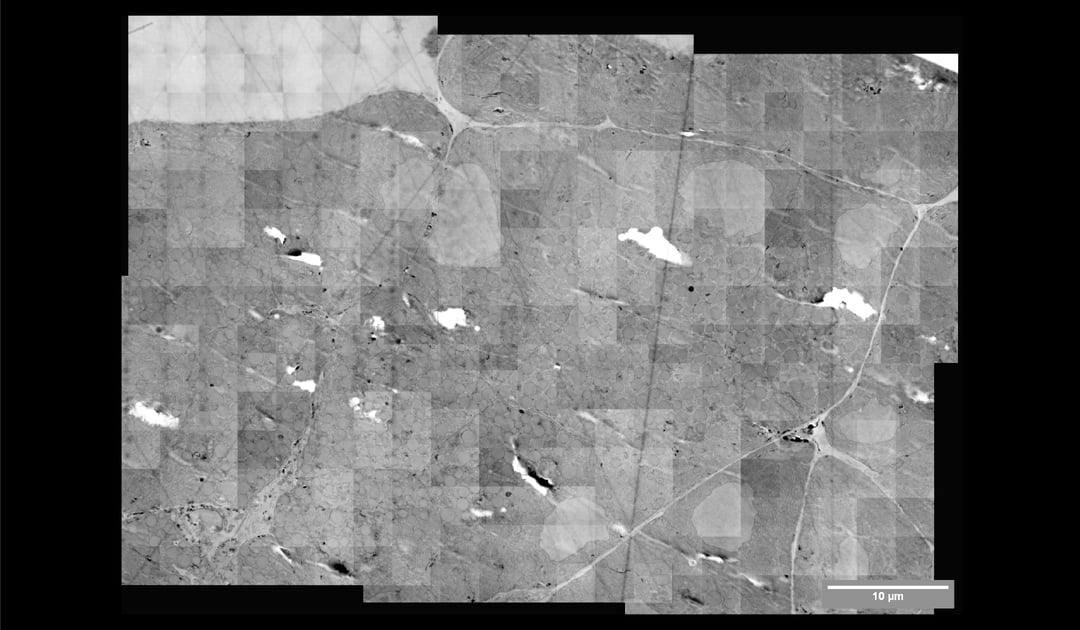
Above you can see the stitched micrographs acquired on Delmic's FAST-EM development prototype. The prototype image shows a region of osmium tetroxide stained, resin-embedded rat pancreas imaged using a prototype of Delmic's novel transmission detector.
References
[1]. E. Sokol et al., “Large-scale electron microscopy maps of patient skin and mucosa provide insight into pathogenesis of blistering diseases,” Journal of Investigative Dermatology, vol. 135, no. 7, pp. 1763–1770, Jul. 2015, doi: 10.1038/jid.2015.109.
[2] A. J. Bushby, G. Mariggi, H. E. J. Armer, and L. M. Collinson, “Correlative light and volume electron microscopy: using focused ion beam scanning electron microscopy to image transient events in model organisms.,” Methods in cell biology, vol. 111, pp. 357–82, Jan. 2012, doi: 10.1016/B978-0-12-416026-2.00018-2.
[3] H. M. Vergara et al., “Whole-body integration of gene expression and single-cell morphology,” bioRxiv, p. 2020.02.26.961037, Feb. 2020, doi: 10.1101/2020.02.26.961037.
[4] Z. Zheng et al., “A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster,” Cell, vol. 174, no. 3, pp. 730-743.e22, 2018, doi: 10.1016/j.cell.2018.06.019.
.png)