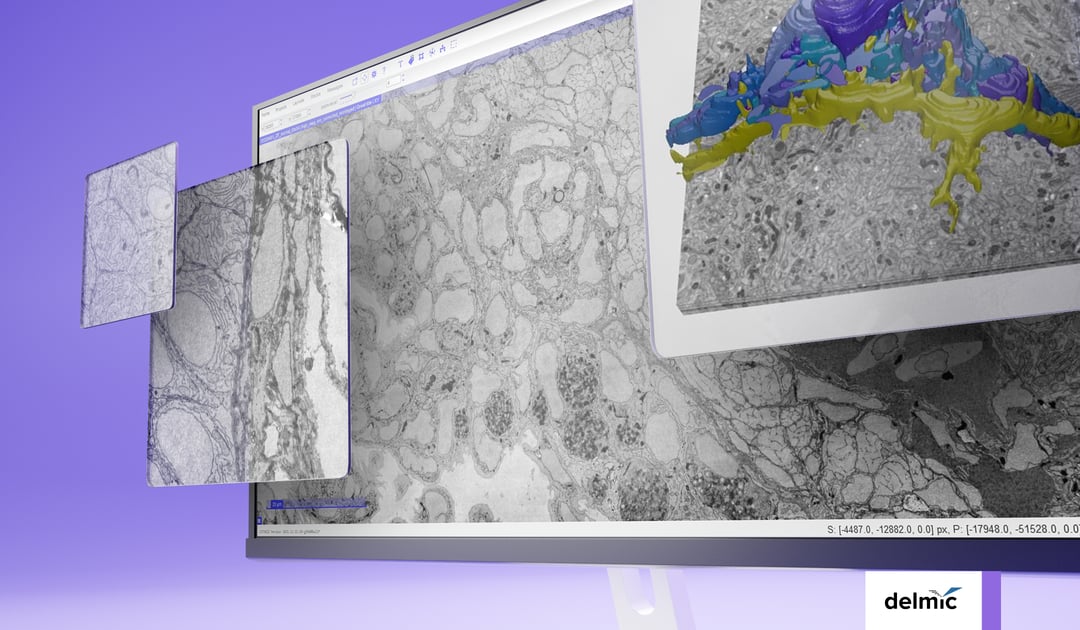
Volume electron microscopy or volume-EM (vEM), acknowledged by Nature as one of the seven technologies to watch in 2023 [1], is increasingly used in life sciences research. It represents a set of techniques and includes the use of either a transmission electron microscope (TEM) or a scanning electron microscope (SEM) [2].
vEM allows organelles, cells, and entire tissue samples to be imaged in three dimensions while achieving nanoscale resolution. The capability to image these large volumes enables researchers to observe and understand the sub-organelle structural details of macromolecular complexes, as well as their tissue context. Therefore, it has already been used in various fields such as connectomics, cancer biology, and virology [3][4][5].
Nonetheless, several challenges stand in the way of a more widespread implementation of vEM techniques in life sciences. The primary obstacle lies in the low throughput of the current vEM workflow, making large-volume imaging difficult and time-consuming [2].
Time-consuming and manual work
The majority of studies use one of three volume electron microscopy techniques: serial block-face SEM (SBF-SEM), focused ion beam SEM (FIB-SEM), or array tomography (AT) [2]. An important step in preparing samples for these techniques involves embedding them in resin and either cutting them into small, thin sections suitable for SEM imaging (AT) or peeling away the top section to reveal a fresh layer of the sample to be imaged (SBF-SEM and FIB-SEM). These processes are very delicate and time-consuming, especially when dealing with a large sample volume.
During SEM imaging, the 2D surfaces of the sample are scanned one at a time. This becomes particularly demanding for larger volumes, which often consist of thousands of these 2D layers. Traditional SEMs operate at a slow pace, as they scan each surface using a single electron beam (Figure 1). Consequently, the complete imaging process for an entire sample can take months.
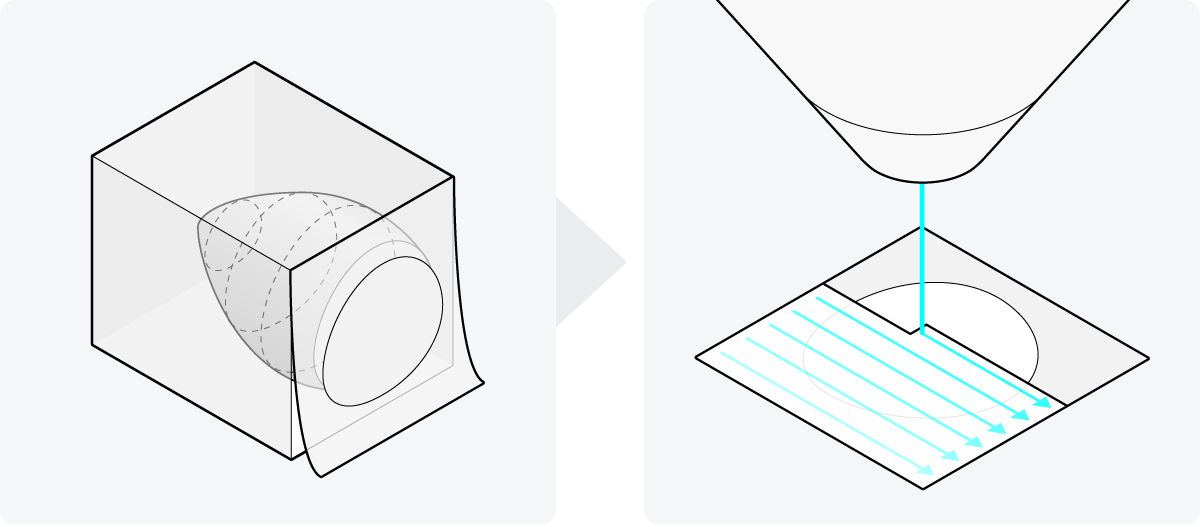
Figure 1: Two steps in vEM sample preparation. The sample is embedded in resin, and cut into small sections or the top section is peeled away in succession. These 2D surfaces are then traditionally scanned with a single electron beam, making the imaging process of a sample extremely lengthy.
Following image acquisition, researchers have often collected more than a terabyte of 2D image data. This data then needs to be transported, as the computer at the electron microscope typically cannot directly display such a large amount of data on its screen. The common method of data transportation involves using portable hard drives, which is impractical for such a large amount of data. Furthermore, these large datasets can’t always be displayed in visualization tools because the whole dataset has to be loaded into the memory. So very costly high-performance computing (HPC) is needed to manage and process the terabyte-sized data.
Apart from the challenges of data visualization, the downstream analysis of the data is also a complex and time-consuming process in vEM, as it’s mostly done manually. There are 3D alignment plug-ins available, but they still require proofreading and correction [6]. As a result, considerable parts of the generated data often remain unanalyzed [5].
Automating sample preparation
Fortunately, significant progress has been made to overcome these bottlenecks. Recent developments include the development of automated section collection techniques for AT to make sample preparation easier and faster [7][8].
Additionally, artificial intelligence (AI) and machine learning (ML) algorithms are being developed to automate various steps in data analysis for vEM. These encompass algorithms for automated stitching, annotation, and segmentation of images [2][9][10]. Furthermore, public archiving of both the vEM data and the implementation of algorithms is of great importance, which is why the EMPIAR public database now supports the collection of vEM data as well (Figure 2) [11].
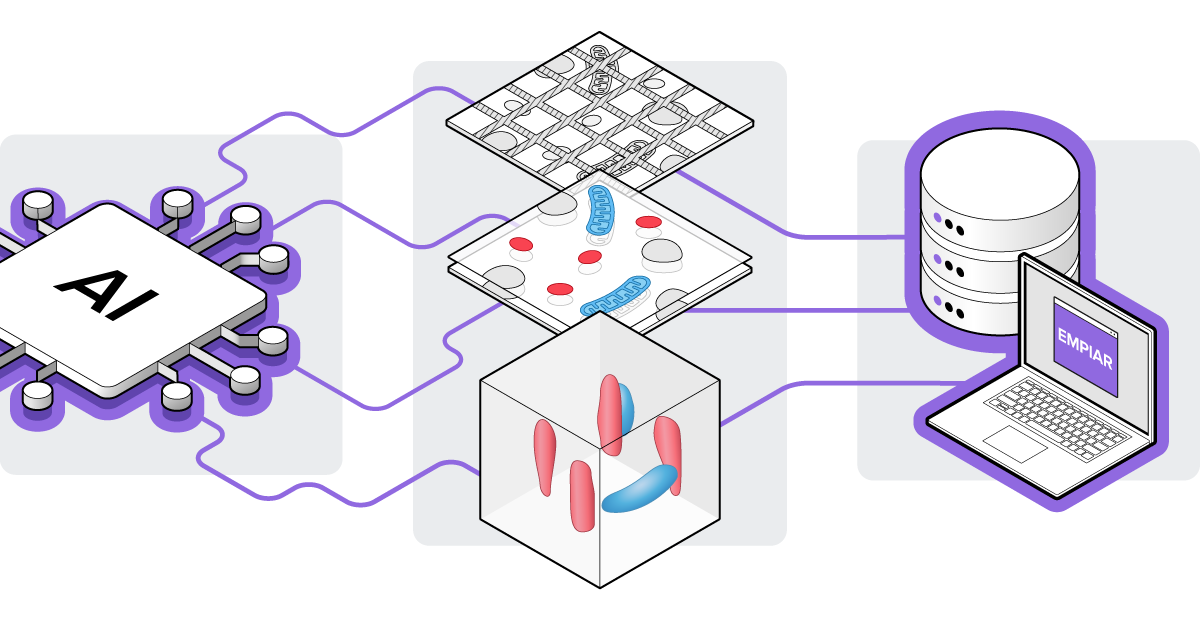 Figure 2: AI and ML algorithms are now being developed that can automate image processes such as alignment and sectioning in vEM datasets. These vEM datasets are nowadays also collected in the EMPIAR public database.
Figure 2: AI and ML algorithms are now being developed that can automate image processes such as alignment and sectioning in vEM datasets. These vEM datasets are nowadays also collected in the EMPIAR public database.
While these solutions help a great deal, in the end, only the electron microscope can substantially change the imaging experience. The electron microscope should not only support these automated solutions but also improve the imaging throughput in the vEM workflow. Only the development of an electron microscope that works fast and allows automation of processes, coupled with the ability to image the samples with adequate resolution, can lead to a more streamlined vEM workflow that is accessible to, and scalable for, life science researchers.
Bringing expertise together
Therefore, Delmic, TU-Delft, Technolution, and Thermo Fisher Scientific worked together to develop an electron microscope that makes the EM workflow as easy and fast as possible while being able to image large areas with nanoscale resolution - The FAST-EM.
To increase the SEM imaging speed for a faster volume-electron microscopy workflow, researchers of the TU-Delft first looked into building multiple electron beams into an existing SEM of Thermo Fisher Scientific. Over a few PhDs, they discovered that the integration of multiple beams in an SEM requires the use of optical components to detect the signals coming from the different beams [12].
Moreover, the TU Delft researchers found that collecting the transmission electrons below the sample yielded much better contrast compared to collecting the scattered electrons above the sample [13]. Thus, due to the better contrast, optical scanning transmission electron microscopy (OSTEM), i.e. an SEM in which the transmission electrons are detected using optical components, was chosen as the key technology for the multi-beam FAST-EM.
By using the scanning transmission electron microscopy principle, images can be generated from multiple electron beams in parallel with little crosstalk. Additional advantages of this setup are the use of scintillators as substrate, as they are rigid and provide stable and ‘bar-less’ support for imaging, minimizing the creasing of sections and ensuring unobstructed imaging areas in the serial sections (Figure 3).
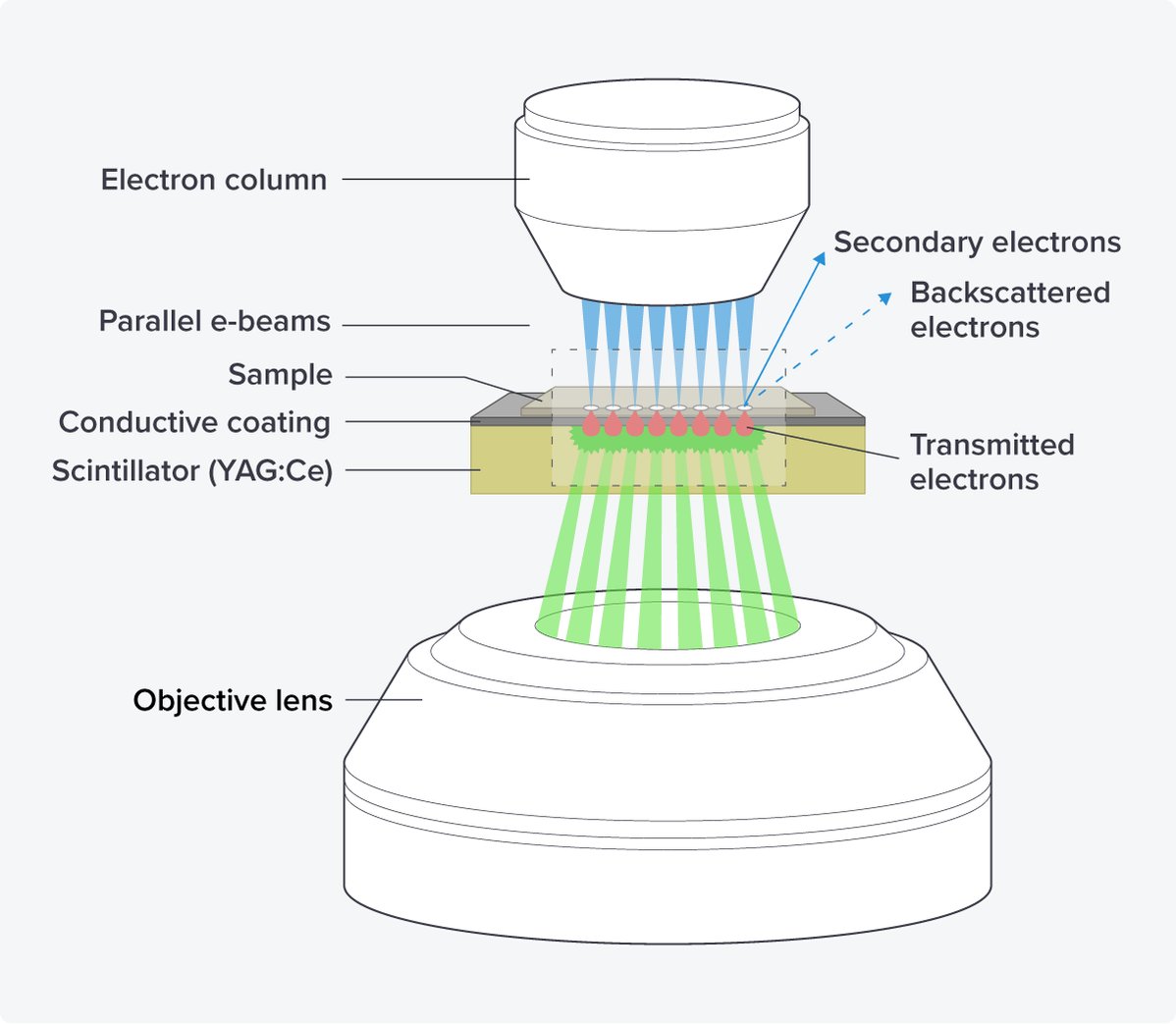
Figure 3: The OSTEM components of the FAST-EM. The electron beams hit the sections of the sample, which are placed on a scintillator with a conductive coating. The transmitted electrons are converted into light in the scintillator, which is subsequently detected by an objective lens.
After the development of the main technique, Delmic, and TU-Delft assessed the optimal number of beams for the commercialization of the system. They found that the use of 64 beams greatly increased the imaging speed (up to 100 times) while achieving 4 nanometer resolution images without crosstalk between the beams [14]. Therefore, the current version of FAST-EM is designed to incorporate 64 scanning electron beams (Figure 4).
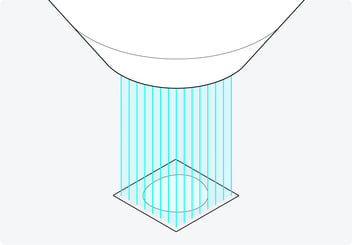
Figure 4: The 64 beams with which the FAST-EM scans the sample in parallel. Using 64 beams greatly increases the imaging speed while achieving high-resolution images without crosstalk.
Less supervision and easy data handling
Then, to make it possible for the researcher to spend less time on the machine while operating, Delmic worked together with Technolution to have the system run on its own without constant supervision. After the sample is positioned under the electron column, brought into focus, and the acquisition parameters are set, automatic beam alignment is ensured using a diagnostics camera and autofocusing.
Lastly, it should be easier for the researcher to handle large datasets and transport them to be able to do data analysis. Technolution has therefore built a dedicated data management platform. Immediately after imaging, the platform automatically stitches together adjacent fields of view. Notably, the platform enables seamless streaming of data from the storage directly to visualization tools such as CATMAID [10] and WebKnossos [15]. Using the CATMAID and WebKnossos browsers, users can navigate the images, zoom in and out, and perform annotation, through an internet connection, so without needing to download the data to their computers (Figure 5).
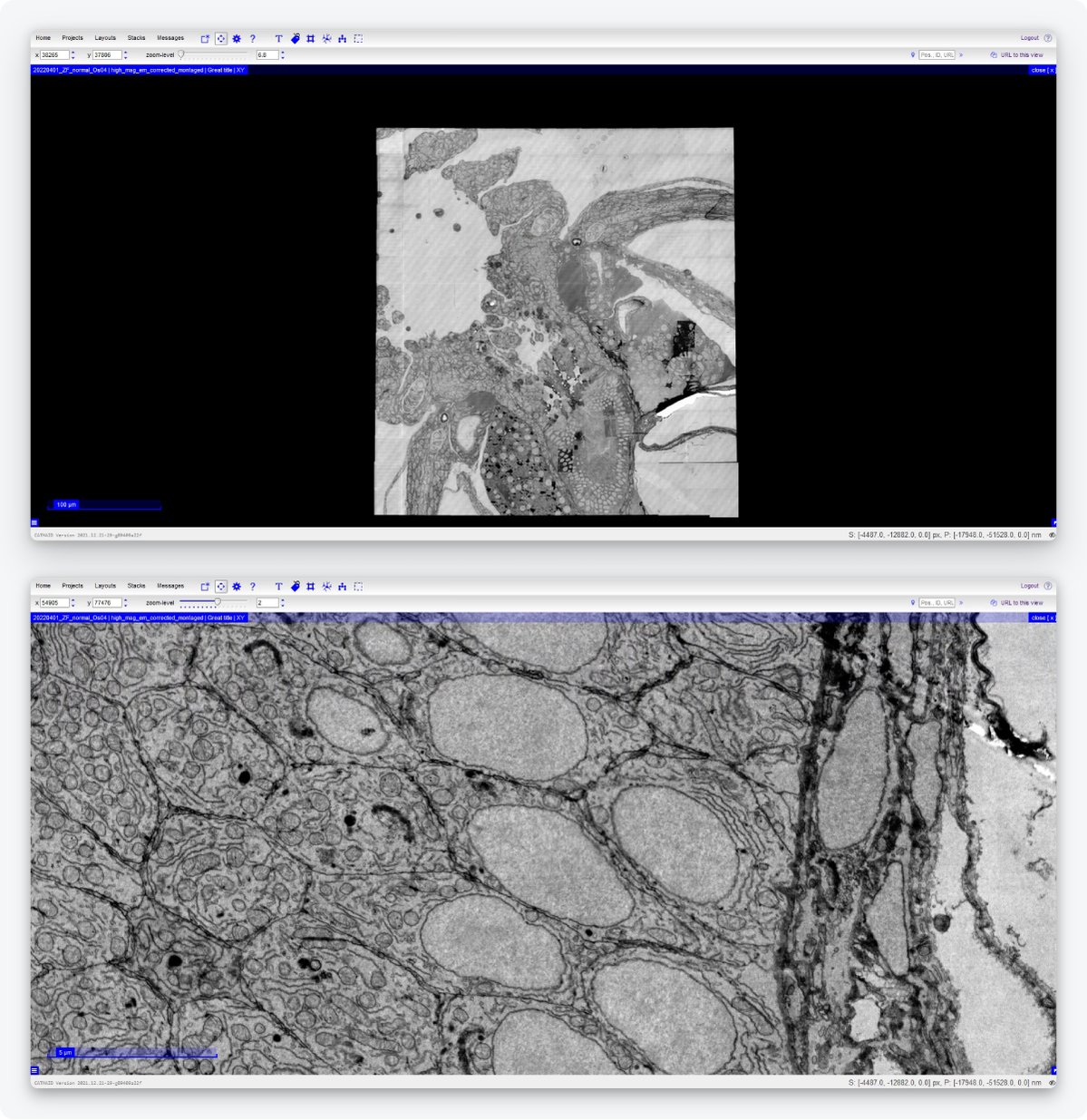
Figure 5: Example of an image acquired by the FAST-EM and displayed on CATMAID. Users can navigate the images, zoom in and out, and perform annotation without having to download the data. Samples made by Ben Giepmans group (UMCG), data acquired by Marre Niesen and Arent Kievits (TU Delft).
So, by bringing various experts together, the FAST-EM was developed. It accelerates image acquisition, works mostly independently, and simplifies data handling and data analysis. The aim is to create a seamless process: the researchers prepare the samples, insert them in the EM chamber, and obtain images for further analysis. Then they can spend their time answering their research questions and publishing their results.
If you had the FAST-EM, how would you spend your gained time?
References
[1] Eisenstein, M., Nature 613, 794-797 (2023)
[2] Kievits, A. J. et al., Journal of Microscopy, 287, 114–137 (2022)
[3] Jessica, L. et al., Methods in Cell Biology 158, 163-181 (2020)
[4] Baena, V. et al., Viruses 13(4), 611 (2021)
[5] Collinson, L.M. et al., Nat Methods 20, 777–782 (2023)
[6] Saalfeld, S., Methods in Cell Biology, Academic Press, 152, 261-276 (2019)
[7] ARTOS 3D ultramicrotome (Leica)
[8] Li, X. et al., Journal of Structural Biology, 200(2), 87-96 (2017)
[9] Siegmund et al.,iScience 6, 83-91 (2018)
[10] Saalfeld, S. et al., Bioinformatics, 25(15), 1984-1986 (2009)
[11] EMPIAR public image archive
[12] Kruit, P. et al., Microscopy and Microanalysis, 25(S2), 1034-1035 (2019)
[13] Zuidema, W. et al., Ultramicroscopy 218, 113055 (2020)
[14] Fermie, J. et al., Microscopy and Microanalysis, 27(S1), 558-560 (2021)
[15] WebKnossos, scalable minds GmbH
.png)