While we understand how most organs function, we still know little about the brain. As brains often consist of billions of neurons that are millimeters in length and even more synapses, understanding how they work is challenging. Researchers in the field of connectomics aim to gain insights into the correlation between the brain structure and its function by building connectomes, which are 3D reconstructions of every neuron and synapse in a certain brain volume.
Visualizing a large area of nanometer-sized synapses is challenging since a high-resolution imaging technique is necessary that can also image large cubic volumes. The wavelength of an electron is much smaller than that of light, so using electron microscopy as an imaging technique is an obvious choice.
For building 3D connectomes in high resolution, volume electron microscopy (volume-EM) is therefore often used [1]. In volume EM, a sample is embedded in resin, and cut into small sections, and these sections are individually imaged under an electron microscope. After image collection and post-processing steps including segmentation, a 3D connectome can be constructed.
Volume-EM is used for mapping increasingly larger brain volumes.
To image large brain volumes in nanoscale resolution, a large amount of data has to be acquired and analyzed. This brings about challenges in both image acquisition and data analysis. Therefore only parts of brains or small invertebrate brains could be reconstructed until recently.
In 2014, researchers investigated how neurons drive motor output from visual input by constructing a connectome of the visual circuit of a Platynereis larva, a marine annelid worm [2]. This animal was chosen because of its small and simple visual system. They used serial sectioning transmission electron microscopy (ssTEM), a volume electron microscopy technique. This allowed them to reconstruct a connectome including photoreceptors, effector muscles, and ciliated cells (Video 1). From the reconstruction, they obtained insights into how the neuronal circuit works in this species and how it might have evolved.
Video 1: Volume rendering of the left primary optic neurophil of a Platynereis larva. The photoreceptor axons (blue) can be seen to form synapses on an interneuron axon (yellow). Two glial cells can be seen in light blue and pink. The positions of the synapses to the axon are shown in different colors for the different photoreceptors. Taken from [2].
Because of advances in data analysis, proofreading, and handling of large datasets, reconstruction of larger brain volumes becomes more achievable [1]. In 2017, researchers reconstructed the connectome of the complete larval zebrafish brain using serial-section electron microscopy [3]. They found remarkable bilateral symmetry of the circuit and now aim to fully understand the structure-function relationship of the neuronal circuit using this large dataset.
Then in 2018, Zheng et al. obtained a connectome of a whole adult Drosophila melanogaster, which was the largest synaptic-resolution dataset at the time [4]. They imaged around 7000 sections and obtained a 106 TB dataset. They could use this dataset to study the mushroom body, which plays a role in learning and even found a previously unknown cell type (Figure 1). The dataset is publicly available to the scientific community.
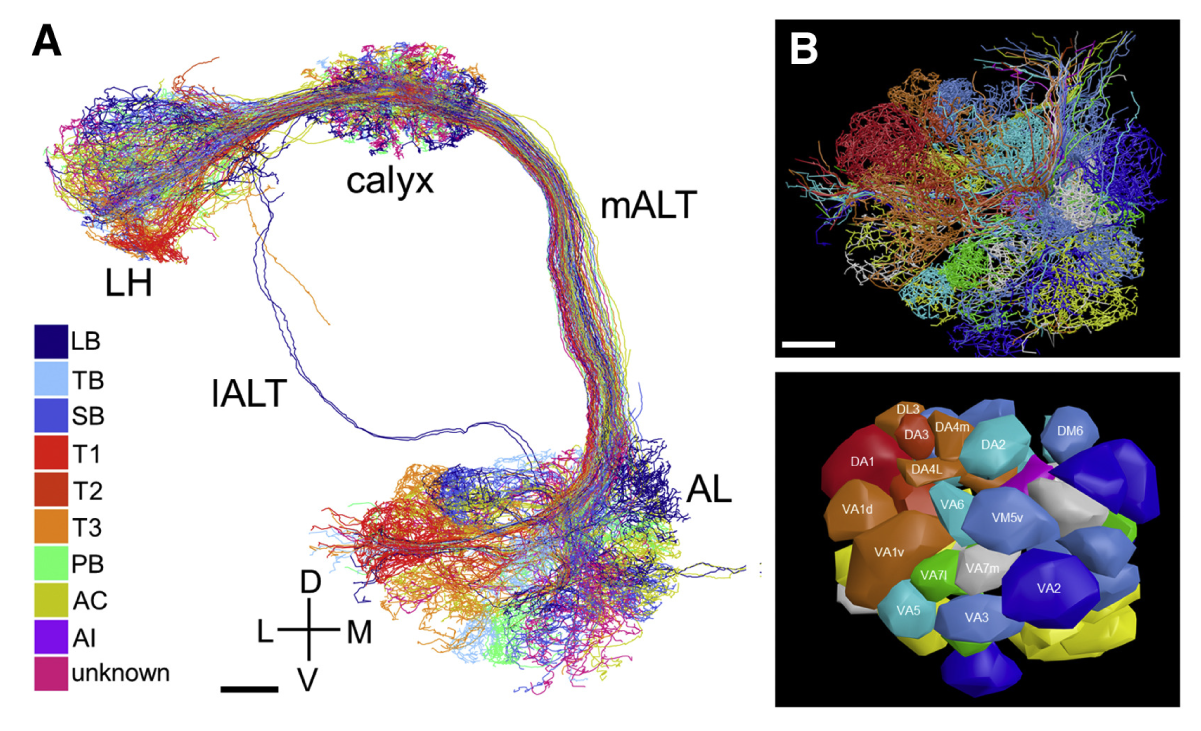
Figure 1: A) Volume-EM reconstruction of uniglomerular olfactory projection neurons in the right hemisphere of a Drosophila brain. B) A frontal view of the AL area, and a glomerular surface model of this area. LH, calyx, mALT, IALT, and AL: anatomical labels in the Drosophila brain. Labeled colors: projection neuron subtypes. Taken from [4].
Recently, researchers performed the largest reconstruction of a mammalian brain to date, namely the neocortex of a mouse [5].
They used volume-EM, and it took them 5 years to complete the dataset. They managed to map 200,000 brain cells and almost 500 million synapses in one cubic millimeter, spanning multiple visual areas of the mouse. They released the dataset for open access, which can be used to drive future research on the mammalian brain.
How can the field of connectomics move forward?
These various studies show that it is possible to use volume-EM for large-scale high-resolution imaging of brains. The main bottlenecks are now mostly the image acquisition speed and the lack of automation in the workflow, making it a laborious imaging method.
Because of these bottlenecks, Delmic has developed the innovative FAST-EM. This scanning transmission electron microscope can perform volume EM imaging up to 100 times faster than a regular scanning electron microscope. The FAST-EM enables imaging without the need for constant supervision. It furthermore helps manage large data files with its innovative data management, which allows for streaming of only the subsets of the data that is in use. In this way, a high-end desktop computer can handle downstream neuron segmentation and analysis. FAST-EM thus makes volume electron microscopy faster and more automated, so that it becomes more accessible and user-friendly for the field of connectomics.
To conclude, there is a clear trend towards imaging large brain volumes at the nanoscale to be able to better understand this complex organ. Advancements in data analysis and image acquisition are crucial for building these large high-resolution reconstructions. Delmic’s innovative FAST-EM could allow for the field of connectomics to grow further, and could thus bring the brain one step closer to understanding itself.
References
[1] Kornfeld et al., Current Opinion in Neurobiology 50, 261-267, (2018)
[2] Randel et al. eLife 3:e02730, (2014).
[3] Hildebrand et al., Nature 545, 345–349 (2017).
[4] Zhihao Zheng et al., Cell 174, 3, 730-743, (2018).
[5] Bae et al., bioRxiv 454025, (2021).
.png)