Throughput of a workflow is often defined as the peak acquisition rate at which a microscope collects image data (usually expressed as pixels per second). However, this peak acquisition rate is rarely maintained throughout a large-scale EM project, since the microscope only spends a portion of its time collecting actual data. Actions like sample exchange, defining regions of interest, or quality control can require a substantial amount of machine time. A more realistic way of looking at a workflow is therefore by defining its sustained throughput, which considers all steps required to collect the hundreds or thousands of images that make up a large-scale EM project. Here, we have a look at the common steps in large-scale imaging, and explore improvements to raise the sustained throughput of a workflow.
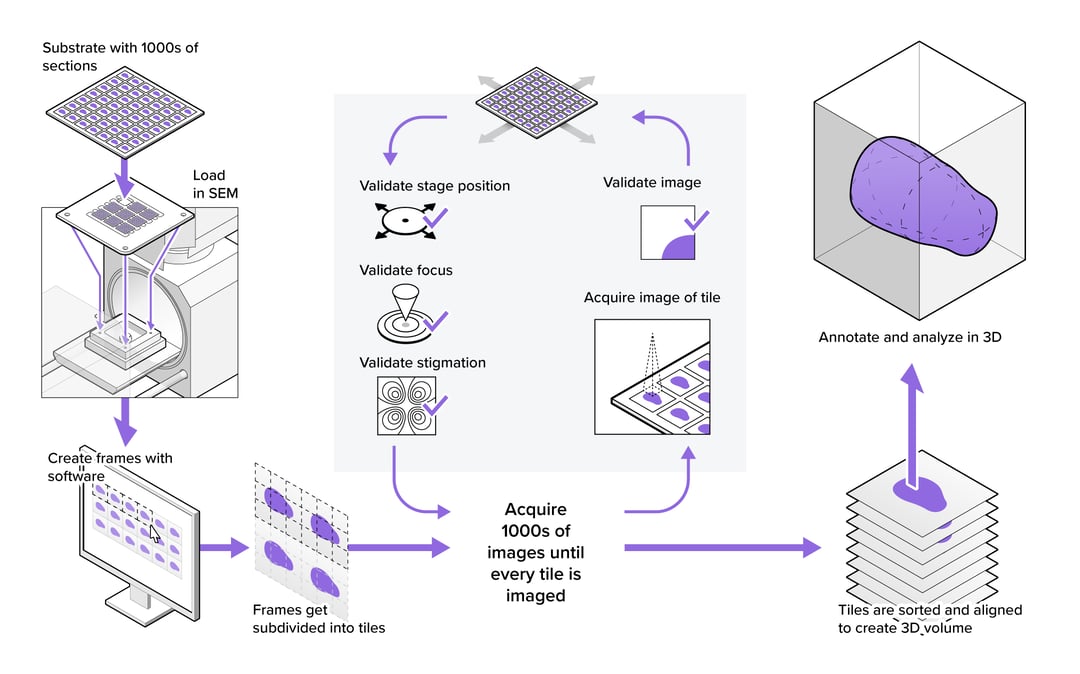
When looking at the overview of a large-scale EM workflow (figure 1), we find that optimizing for throughput can start with basic steps like sample exchange. In many cases, only a fraction of samples in the project can be loaded in the microscope simultaneously, so samples need to be exchanged regularly. Sample exchange is time-consuming and disrupts the vacuum quality of the EM, so it is beneficial to minimize the time required per sample exchange and minimize the number of sample exchanges needed. The former can be achieved by automating the loading and unloading of samples (1), whereas the latter can be accomplished by maximizing the number of sections loaded at once with larger or continuous specimen holders (2–4).
A second area where leaps can be made is the need for user input, which comes up at multiple stages in the projects. First, operators need to specify project details on the microscope. Significant time may be needed to set up a project exactly to the demands of the operator, during which the microscope is not collecting usable data. Once the system is collecting data, the challenge shifts to minimizing the interaction required to keep the system running consistently. The robustness of the hardware and software can make the difference between a microscope requiring constant babysitting or running unattended for days or weeks at a time.
Finally, there is the ‘imaging cycle’ which is used to automatically capture all images in the project, and which may be the most important target to increase sustained throughput (figure 1). Most large-scale workflows automate actions to ensure the quality of every image, which includes stage movement, focusing, stigmation, and artefact detection. These are an essential, but potentially time-consuming part of the imaging cycle. Each of these actions is repeated thousands of times over the course of a project, depending on how stable the microscope is. Even small improvements to their efficiency can therefore improve the throughput of the imaging cycle substantially.
In conclusion, building a workflow with high sustained throughput goes beyond merely achieving a high peak acquisition rate. It requires a holistic view of the workflow, where all actions in the process are considered and optimized. Delmic is currently working on a high-speed electron microscopy solution to examine large samples with a high sustained throughput. With this solution, we will provide a stable, automated workflow based around minimal operator intervention.
If you would like to learn more about the possibilities of fast electron microscopy and Delmic's solution, watch our on-demand webinar Fast Electron Microscopy for large-scale projects.
Figure 1: Example of a large-scale electron microscopy workflow for volume EM, showing the preparatory steps before imaging, the imaging cycle required to acquire all tiles in the dataset, and the offloading and processing of the image data.
References
[1] Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, et al. A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell [Internet]. 2018;174(3):730-743.e22. Available from: https://doi.org/10.1016/j.cell.2018.06.019
[2] Horstmann H, Körber C, Sätzler K, Aydin D, Kuner T. Serial section scanning electron microscopy (S 3EM) on silicon wafers for ultra-structural volume imaging of cells and tissues. PLoS ONE. 2012;7(4).
[3] Yin W, Brittain D, Borseth J, Scott ME, Williams D, Perkins J, et al. A Petascale Automated Imaging Pipeline for Mapping Neuronal Circuits with High-throughput Transmission Electron Microscopy. bioRxiv [Internet]. 2019 [cited 2020 Feb 4];791889. Available from: https://www.biorxiv.org/content/10.1101/791889v1
[4] Hayworth KJ, Morgan JL, Schalek R, Berger DR, Hildebrand DGC, Lichtman JW. Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Frontiers in Neural Circuits [Internet]. 2014;8(June):1–18. Available from: http://journal.frontiersin.org/article/10.3389/fncir.2014.00068/abstract
.png)