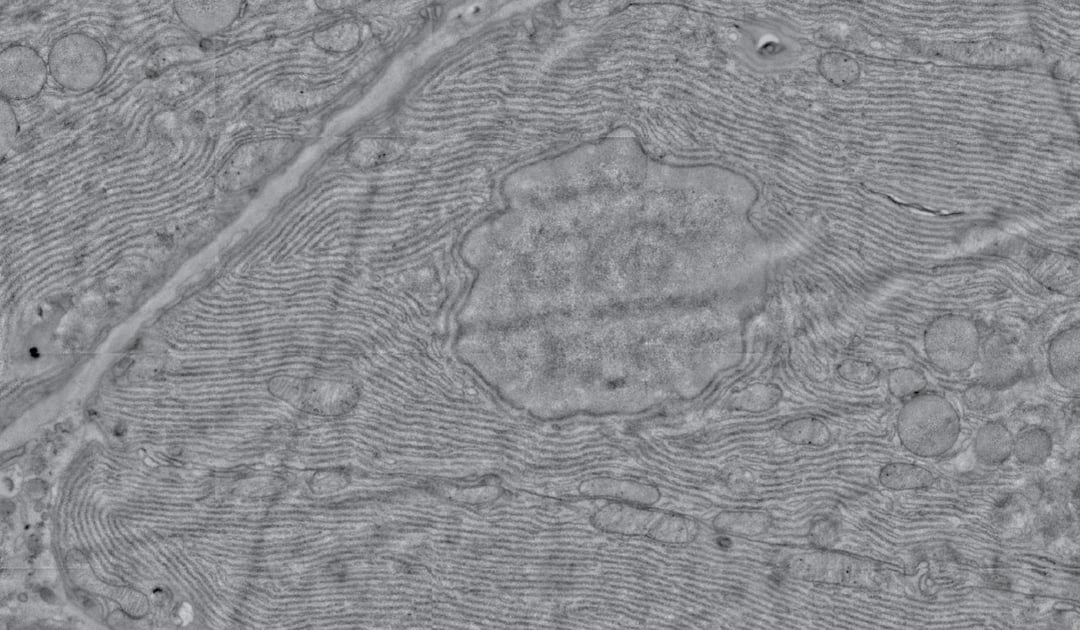
Electron microscopy (EM) is increasingly being used in large-scale biological projects, either for volume imaging or large area mapping. In both cases, the desire to resolve nanoscale details is combined with the desire to place these observations into a larger context, which can cover a large area or span through a three-dimensional volume. Thanks to increased automation and image processing efforts, these large-scale imaging projects are now feasible. Information from these projects has already proven highly beneficial for a variety of research fields, including developmental and cell biology, neurosciences and pathology [1, 2, 3].
Still, large-scale imaging presents an interesting situation, where nanoscale resolution is needed over an area or volume that is several orders of magnitude larger than the field of view of the microscope. This provides several major challenges for conventional scanning EMs. Watch our discussion about major bottlenecks of EM.
One of the biggest challenges is the limited speed of current scanning EM systems. Due to their limited acquisition rate microscope operators are forced to balance the size of the imaged area against the resolution needed to visualize the required level of detail. Despite best efforts, large projects still take days to months of continuous imaging on multiple machines when high-resolution data is needed [4, 5, 6]. This produces a logistical issue for imaging facilities, which only have limited capability of handling these projects for more than a select few users.
Another challenge in large-scale imaging is reducing overhead in the imaging cycle, which affects the sustained throughput of a microscope. Throughput is often advertised as the peak acquisition rate of the microscope (usually expressed as pixels per second). However, this peak acquisition rate is seldom achieved during an imaging project, thanks to overhead in the imaging cycle caused by sample exchange, stage movement and image quality adjustment. These steps in the imaging cycle quickly amount to at least 30-40% of the microscope time [3, 7, 8]. Efficient acquisition routines that minimize microscope overhead are therefore crucial to maximize the sustained throughput of any EM workflow.
Finally, conventional microscopes are not yet automated enough to function unattended for large projects. Some form of region selection is necessary for large-scale applications, which is usually done manually by an operator depending on the question or project. In addition, conventional microscopes often do not run extended periods of time without an operator babysitting the system to handle tasks like sample exchange and selection of imaging regions, beam focus and alignment, limiting their potential for extended imaging runs. In the worst case, this means the microscope only produces data during working hours of operators, which significantly impacts throughput.
The need for increased throughput, reliability and automation of electron microscopy reveals the need for a drastic new approach to large-scale imaging. Delmic's automated fast scanning electron microscope FAST-EM is the solution to overcome the challenges that slow down large-scale electron imaging. Currently Delmic is working in collaboration with Thermo Fisher, TU Delft and Technolution on a highly automated and reliable workflow, which will be accessible to a broad set of users due to its ease of use.
In the upcoming months we will be highlighting other aspects of fast EM imaging, and the project we are currently working on. Would you like to be updated and be the first one to know about the system we are working on? Please click the button below.
References:
[1] Peddie, C. J., & Collinson, L. M. (2014). Exploring the third dimension: volume electron microscopy comes of age. Micron (Oxford, England : 1993), 61, 9–19. https://doi.org/10.1016/j.micron.2014.01.009
[2] Sokol, E., Kramer, D., Diercks, G. F. H., Kuipers, J., Jonkman, M. F., Pas, H. H., & Giepmans, B. N. G. (2015). Large-scale electron microscopy maps of patient skin and mucosa provide insight into pathogenesis of blistering diseases. Journal of Investigative Dermatology, 135(7), 1763–1770. https://doi.org/10.1038/jid.2015.109
[3] Titze, B., & Genoud, C. (2016). Volume scanning electron microscopy for imaging biological ultrastructure. Biology of the Cell, 108(11), 307–323. https://doi.org/10.1111/boc.201600024
[4] Hildebrand, D. G. C., Cicconet, M., Torres, R. M., Choi, W., Quan, T. M., Moon, J., Wetzel, A. W., Scott Champion, A., Graham, B. J., Randlett, O., Plummer, G. S., Portugues, R., Bianco, I. H., Saalfeld, S., Baden, A. D., Lillaney, K., Burns, R., Vogelstein, J. T., Schier, A. F., … Engert, F. (2017). Whole-brain serial-section electron microscopy in larval zebrafish. Nature, 545(7654), 345–349. https://doi.org/10.1038/nature22356
[5] Xu, C. S., Hayworth, K. J., Lu, Z., Grob, P., Hassan, A. M., García-Cerdán, J. G., Niyogi, K. K., Nogales, E., Weinberg, R. J., & Hess, H. F. (2017). Enhanced FIB-SEM systems for large-volume 3D imaging. ELife, 6, 1–36. https://doi.org/10.7554/eLife.25916
[6] Yin, W., Brittain, D., Borseth, J., Scott, M. E., Williams, D., Perkins, J., Own, C., Murfitt, M., Torres, R. M., Kapner, D., Bleckert, A., Castelli, D., Reid, D., Lee, W.-C. A., Graham, B. J., Takeno, M., Bumbarger, D. J., Farrell, C., Reid, R. C., & Costa, N. M. da. (2019). A Petascale Automated Imaging Pipeline for Mapping Neuronal Circuits with High-throughput Transmission Electron Microscopy. BioRxiv, 791889. https://doi.org/10.1101/791889
[7] Briggman, K. L., & Bock, D. D. (2012). Volume electron microscopy for neuronal circuit reconstruction. Current Opinion in Neurobiology, 22(1), 154–161. https://doi.org/10.1016/j.conb.2011.10.022
[8] Zheng, Z., Lauritzen, J. S., Perlman, E., Robinson, C. G., Nichols, M., Milkie, D., Torrens, O., Price, J., Fisher, C. B., Sharifi, N., Calle-Schuler, S. A., Kmecova, L., Ali, I. J., Karsh, B., Trautman, E. T., Bogovic, J. A., Hanslovsky, P., Jefferis, G. S. X. E., Kazhdan, M., … Bock, D. D. (2018). A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell, 174(3), 730-743.e22. https://doi.org/10.1016/j.cell.2018.06.019
.png)