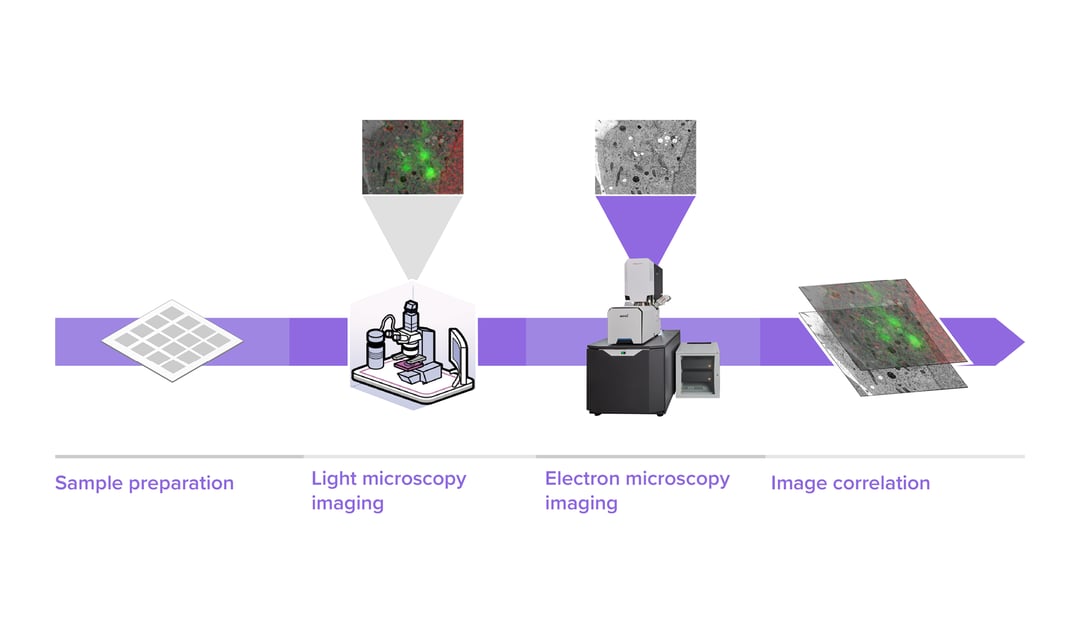
In order to analyse various aspects of the complex organisation of cells, there is increasing demand to study the same sample at different length scales in biology. Correlative light and electron microscopy (CLEM) is a powerful technique that is used for this purpose. CLEM combines fluorescence microscopy (FM) with high-resolution electron microscopy (EM) to provide information about the composition and function of the sample.
Light microscopy (LM) is extensively used as a screening tool for morphological hallmarks between individuals or in response to drugs and genetic alterations. Its high throughput and large field of view enable rapid data collection for different experimental conditions. Electron microscopy (EM), on the other hand, is a powerful technique for examining membrane structures and ultrastructure in general. Combining these two techniques could provide the benefits of understanding the structure underlying changed behavior or composition of cells and organelles, as observed by FM.
CLEM workflows usually start by looking at thin sections of a sample with fluorescence microscopy, choosing the region of interest (ROI) and then transferring the section to an SEM or TEM for further investigation. The next step of the process is to collect the images of the selected regions with electron microscopy, which can be extremely time-consuming if larger areas are required at high resolution. For this reason, cell biologists may only able to retrieve information on a few single organelles at a very high resolution, without acquiring high-resolution images of the surrounding areas. Instead, a low resolution of the whole field of view is made to check orientation, followed by imaging a small number of the most relevant labels.
In such traditional CLEM workflows, EM is a limiting factor for the scope of data collected: with low throughput and limited field of view, careful selection of ROIs is needed to ensure the feasibility and success of a project. Moreover, the limited throughput of the workflow leads to the lack of context, which makes CLEM more of a qualitative technique rather than quantitative.
The techniques used for electron microscopy have continuously improved over the years, making it possible to match the scale of fluorescence imaging with electron microscopy techniques, while retaining the resolution required to visualise the fine details of the cell. However, this still means that collecting large-scale data requires a long time with a conventional electron microscope.
With FAST-EM, CLEM workflows can be enormously simplified and can easily provide quantitative information to the researchers. Indeed, FAST-EM is a multibeam electron microscope capable of imaging the samples 100 times faster. A megafield of the dimensions of roughly 250 µm x 250 µm, with 4 nm pixel size, can be collected within just 3 minutes. The system allows observing fine morphological details while retaining the overview and the context. Moreover, FAST-EM substrates offer a large, uninterrupted sample surface, while the technique behind FAST-EM is compatible with mildly-stained samples.
Combined with high-throughput EM offered by FAST-EM, larger volumes can be investigated by CLEM, enabling a transformation towards integration of imaging modalities on a large scale. Instead of gathering data only on the few labelled regions of interest, it is possible to capture the whole field of view with EM and correlate it with the FM data. Such large-scale acquisition enables morphometric analysis with EM [1]. Having information on many labelled regions, it is possible to compare the sizes and shapes of organelles between cell types or different experimental conditions. This type of quantitative analysis typically requires weeks or months to be completed with standard single-beam systems. With FAST-EM it could be completed within a few hours or days.
To learn more about the system make sure to join our upcoming FAST-EM webinar.
References
[1] J. M. Serra Lleti et al., "CLEMSite, a software for automated phenotypic screens using light microscopy and FIB-SEM," BioRxiv, doi: https://doi.org/10.1101/2021.03.19.436113
.png)