Electron microscopy (EM) is increasingly used in the field of life sciences. It can be used to study proteins, macromolecules, and tissues both in vitro and in situ. Since life is three-dimensional and contains nanoscale details, imaging large in situ samples in 3D using EM can provide a precise depiction of life.
The demand for 3D imaging of increasingly larger samples, combined with technological advancements, has led to the emergence of volume electron microscopy (volume-EM). Volume-EM represents a set of techniques that allow sample volumes to be imaged in 3D at the nanoscale. It provides high-resolution images of organelles within their 3D tissue context. This gives a more accurate depiction of e.g. the inner workings of cells in healthy and diseased tissue. Notably, Nature highlighted volume-EM as one of the ‘seven technologies to watch in 2023’ [1].
Despite its huge potential, the volume-EM community is still in its early stages, and several issues need to be addressed to make it more widely available and applicable. So what are these limitations, and how can we overcome them?
Accelerated sample preparation
In the electron microscopy workflow, biological samples must be preserved in their natural state while being bombarded with electrons in a vacuum chamber. This can be achieved through various sample preparation steps, which are optimized and vary depending on the sample. In general, the sample preparation process involves 1) optimization of the sample size, 2) fixing the sample, 3) staining with contrast agents, 4) dehydrating the sample and embedding it into a resin, and 5) trimming the resin block.
For sample fixation, the methods used fall into two main categories: cryofixation and chemical fixation. Cryofixation is widely used for fixing protein structures and macromolecules in their native state, but it has a limited depth of vitrification of only 200 μm, making it unsuitable for imaging large volumes. While chemically fixed sections may contain shrinkage artifacts, it has been found to perform equally well as cryo-EM does for imaging large volumes [2].
After contrast staining, dehydration, and resin embedding, the sample has to be cut into small thin sections. With the emergence of the ultramicrotome, this process has already been improved in terms of speed, z-resolution, and the ability to re-image sections if necessary (Figure 1) [3]. Additionally, ARTOS 3D (Leica) overcomes the labor-intensive process of manual sectioning by automatically creating and collecting ultrathin sections [4]. By further optimizing the volume-EM sample preparation workflow, and standardizing it per sample type, volume-EM could become a more accessible and less time-consuming technique.
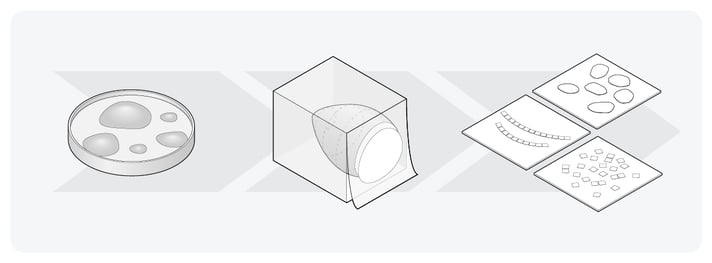 Figure 1: The main steps in the Volume-EM sample preparation workflow. The samples are embedded in resin, and subsequently cut into small thin sections.
Figure 1: The main steps in the Volume-EM sample preparation workflow. The samples are embedded in resin, and subsequently cut into small thin sections.
Faster image acquisition
After preparing the sample sections, images are mainly acquired using either a scanning electron microscope (SEM) or a transmission electron microscope (TEM). However, a tradeoff exists between imaging speed, resolution, and the volume that can be imaged during the imaging process [2]. When using an SEM, imaging is slow because a single SEM beam has to scan the entire sample at a high resolution.
Furthermore, acquiring images is often done manually, which is also very time-consuming [5]. Mapping a part of the brain in the field of connectomics can take years. To overcome these bottlenecks, electron microscopes such as Delmic’s FAST-EM have been developed, which is a multi-beam scanning TEM (STEM) that can image large volumes faster than a regular SEM.
Automated image reconstruction and data analysis
Following image acquisition, the 2D images have to be processed, and a 3D reconstruction has to be made. This is done by aligning the images in a single volume (Figure 2), and subsequently stitching together the smaller volumes. There is ongoing research into using machine learning techniques to optimize and automate the alignment of images [2].
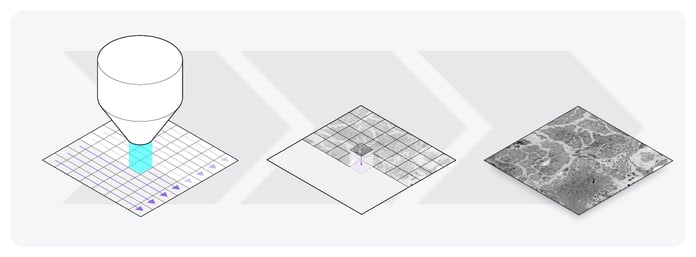 Figure 2: Image acquisition of one section of the sample. First, the electron microscope scans the sample. In this illustration, the sample is scanned using the FAST-EM, which scans with 64 beams instead of one. Then, the small 2D images are aligned, and a large 2D image is reconstructed.
Figure 2: Image acquisition of one section of the sample. First, the electron microscope scans the sample. In this illustration, the sample is scanned using the FAST-EM, which scans with 64 beams instead of one. Then, the small 2D images are aligned, and a large 2D image is reconstructed.
Once the 3D reconstructions are complete, they’re enhanced and analyzed. This requires visualization of the dataset. However, due to the large size of volume-EM datasets (terabytes), visualizing the data is challenging [5]. Researchers often need to transfer the data files from the electron microscope to their computer, since the data files are too large to view on the screen. This requires the researcher to use USB sticks to physically transport the data to high-end computers for analysis. Moreover, many visualization tools are unable to explore large datasets, as often all the data has to be loaded into the memory [2].
To address these limitations, the image acquisition software of the electron microscope should provide support for pre-computed multiscale image pyramid format and chunk-based data access. Additionally, storing the data in the cloud would omit the need to physically transport large data files, saving researchers a lot of time [2]. After uploading to the cloud, researchers can stream the data on platforms such as CATMAID (Figure 3) [6].
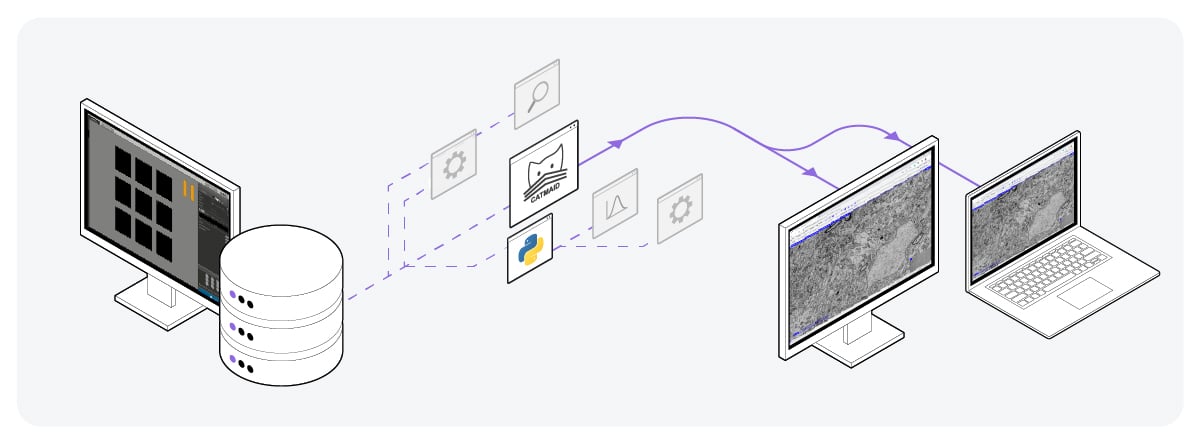 Figure 3: Volume-EM data storage by uploading on the cloud. Using platforms such as CATMAID, the data files can be easily accessed, viewed, and shared from anywhere.
Figure 3: Volume-EM data storage by uploading on the cloud. Using platforms such as CATMAID, the data files can be easily accessed, viewed, and shared from anywhere.
Apart from data visualization, data analysis is also a difficult and time-consuming process in volume-EM, as it’s mostly done manually. Although significant progress has been made in recent years to develop artificial intelligence (AI) algorithms that can perform the segmentation of features in the images, there is not yet a reliable and easy-to-use algorithm that can automate this process [2]. The rapid development of novel AI algorithms and open-source data analysis tools will help overcome this bottleneck in the near future.
The bright future of volume-EM
At Delmic, we firmly believe that the problems of volume-EM can be overcome by enabling faster and more automated usage of the technique, combining cutting-edge technology with AI-based data analysis. We envision volume-EM as an imaging technique that can be used everywhere, to observe the nanoscale easily in three dimensions. Do you want to know more? Watch our video series that explores the complexities and big ideas behind volume EM and most importantly, the future of EM!
References
[1] Eisenstein, M., Nature 613, 794-797 (2023)
[2] Peddie, C.J. et al., Nat. Rev. Methods Primers 2, 51 (2022)
[3] Titze, B. et al., Biol. Cell, 108, 307-323 (2016)
[4] ARTOS 3D website Leica
[5] Collinson, L.M. et al., Nat. Methods (2023)
[6] Saalfeld, S. et al., Bioinformatics, 25(15), 1984–1986 (2009)
.png)