The College of American Pathologists defines pathology as "the medical discipline that provides diagnostic information to patients and clinicians."[1]. Because pathology plays a fundamental role in our understanding of diseases and patient care, including diagnosis, prognosis, and treatment guidance, it is important that the technologies used by pathologists enable them to perform their work as efficiently as possible while delivering the most accurate results.
To visualize abnormal structural changes in cells and tissues, pathologists routinely use brightfield light microscopy (LM). However, the resolution of these techniques typically does not allow them to look at structures smaller than 400 nm. As we advance our understanding of biological processes at the nanoscale, the ability to visualize pathological agents and processes below the resolution of routine light microscopes becomes increasingly important. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM), with their much higher resolving power, are the obvious choices.
TEM was routinely used as a key technique in pathology diagnosis in the 80’s but is replaced by other advanced diagnostic technologies such as immunohistochemistry and Next Gen Sequencing. In some areas of pathology, such as kidney, skin, and muscle pathology, TEM is still a crucial tool for diagnosis because it provides the required nanometer resolution to visualize the ultrastructural changes in these tissues. However, although digital pathology with whole slide imaging has already become a common practice in pathology at the LM stage, digital ultrapathology still seems to be a faraway dream.
TEM imaging at pathology laboratories are at present still operated manually by skilled technicians to select specific areas to image at high magnification, resulting in a limited diagnosis capacity and the human bias. Conventional single-beam SEMs are also limited in imaging speed. These factors limit the sampling throughput and therefore the capacity for TEM-based diagnosis. In addition, data storage needs to be adequate before ‘digital ultrapathology’ can become a reality.
With the introduction of automated multi-beam SEM, such as our FAST-EM, the abovementioned drawbacks can be eliminated, offering the possibilities to unlock the benefits of EM for ultrapathology.
Let’s explore in more detail the use of TEM and the benefits of its next-generation, fast, and automated version combined with AI-based data analysis in two exemplary applications: renal pathology and coronavirus research.
Impact of the high-throughput EM on renal pathology
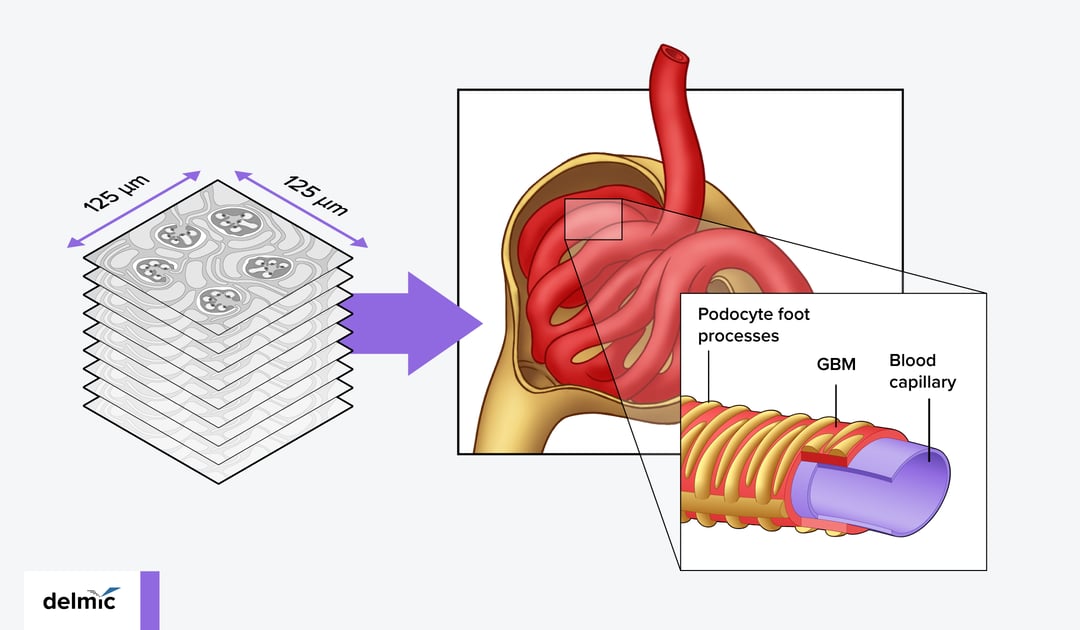
Pathological analyses in several serious kidney diseases focus on the glomeruli, i.e. the structures where the filtration of selected blood components occurs. This ultrafiltration is regulated by three major elements of a glomerulus: endothelial cells, podocytes, and the glomerular basement membrane (GBM) [2] (Figure 1). Structural changes and the resulting malfunctioning of these components lead to potentially life-threatening conditions, such as diabetic kidney disease (DKD). DKD develops in up to 50% of the patients with type 2 diabetes, and up to 30% in those with type 1 diabetes [2, 3]. One of the first manifestations of the development of DKD is a diffuse thickening of the GBM. These changes develop over a span of years, without giving any clinical symptoms [4]. Once the symptoms occur, however, the distortions are so advanced, that the glomerular filtration rate declines rapidly, ultimately leading to kidney failure. That is why the check-ups for any structural changes in the GBM and other glomerular components in the risk group patients are critical [5].
In a traditional TEM-based diagnosis of a kidney disease, a technician takes TEM images of the glomeruli from the resin-embedded sample sections and selects specific locations to acquire high magnification images, which are then shared with a pathologist. After measuring the thickness of the GBM manually at three locations on the image, the technician provides the calculated average to a pathologist for diagnosis. This procedure thus involves several manual steps and subjective decisions that may result in inaccuracies in the pathological analysis. A further drawback of a traditional TEM is the limited sample area that can be investigated at a high resolution [6].
By using an automated multibeam SEM, technicians are able to acquire images across larger areas of the sample at a high resolution and perform volume EM imaging via array tomography (Figure 1). Scanning a larger section at a high resolution lowers the bias that can occur compared with imaging at high magnification at manually selected regions. It also allows the technicians to better account for the heterogeneous nature of tissues. Similar to whole slide imaging in digital pathology, the SEM images can be examined again later if more information is needed. Moreover, artificial intelligence (AI) algorithms may be developed [7] and applied to segment the data and ultimately extract specific parameters (e.g. thickness, density, cell-cell contact) of the particular tissue structure or organelle for quantitative analyses.
Volume imaging can offer further advantages in the case of pathological analysis of kidney diseases, where sample sectioning can introduce bias in the GBM thickness measurement (Figure 2) and the quantification of podocyte contact with the basement membrane. Instead, volume imaging whole glomeruli provides a more accurate assessment of the basement membrane thickness.
Figure 2: Currently the glomerular basement membrane (GBM) thickness is measured based on the EM images recorded from a single section, which can create bias in the measurement depending on the way the patient’s specimen is cut.
Besides being a diagnostics tool, high-throughput SEM could also be applied to better understand the mechanism of drugs used for the treatment of renal diseases, such as metformin, which was shown to have beneficial effects on podocytes and epithelial cells [3].
Impact of the high-throughput EM on coronavirus research
The high resolution of EM techniques makes it possible to visualize not only individual viral particles but even their structural characteristics, such as double membrane vesicle structures typical for Coronavirus virions (Figure 3). That is why, when COVID-19 was spreading across the world, many scientific groups applied their EM systems to reveal the mechanisms of cell infection by SARS-CoV-2 viruses [9,10]. Using SEM and segmentation it is moreover possible to visualize and quantify the different effects the SARS-CoV-2 variants have on the cellular morphology. With a high-throughput SEM, virologists and pathologists can deepen their understanding through these nanoscale details over larger sample areas and investigate more sample types. The resulting quantitative characterizations and comparisons between patients, time points, tissue areas, etc., enable them to better understand disease manifestation, heterogeneity, and progression. AI auto segmentation models could moreover be developed by specialists and applied to discriminate virion features. As a result, a high-throughput SEM can serve as a fast and reliable assessment technique for different SARS-CoV-2 infected samples and models.
Figure 3: high resolution EM can reveal insights pertaining to infection pathways, such as the observation of double membrane vesicles predominantly in ciliated cells rather than goblet cells.
If you are interested to learn how our high-throughput SEM system FAST-EM was applied to SARS-CoV-2, check our application note 'FAST-EM to accelerate and deepen the insight into infectious disease' below.
Until recently, the ability to visualize large sample areas on a nanoscale using SEM/TEM was not practical due to the low throughput and manual nature of these techniques. This has changed with the introduction of multi-beam SEM. Our FAST-EM system images up to 100x faster than conventional SEMs and can be run unsupervised for up to three days. What would take weeks to image on a single beam SEM now takes hours with FAST-EM. FAST-EM comes with an innovative data management platform. Specialists can use the collected data to build AI-based segmentation models that can then be applied to automated data analysis. In the hands of specialists working in pathology and drug development labs, FAST-EM combined with AI data analysis can enable increased diagnostic capacity and accuracy, thereby improved prognosis and facilitate the characterisation of the effects of drug treatments in a more quantitative manner.
References
[1] What is pathology? College of American Pathologists
[2] Kamenetsky et al. , J Digit Imaging, (2010).
[3] Lehtonen S., Pharmaceuticals (Basel), (2020)
[4] Marshall CB., Am J Physiol Renal Physiol, (2016)
[5] Conti et al., Sci Rep 8, (2018)
[6] Masum et al., Sci Rep 8, (2018)
[7] Chengqing et al., BioRxiv preprint, (2021)
[8] Caldas et al., Sci Rep 10, (2020)
[9] Pinto et al., Nat Commun 13, (2022)
[10] Hopfer et al., Histopathology 78, (2021)
.png)