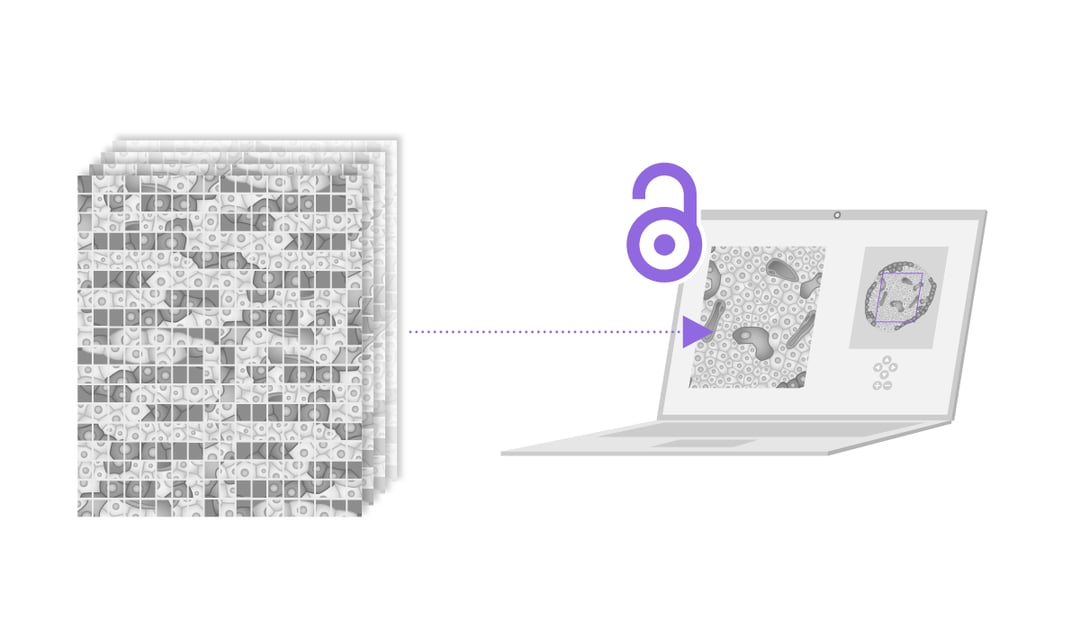
Even though the amount of research done on diabetes is considerable, there is still a large gap in what we know of the pathophysiological mechanisms of diabetes type 1. In this blog post two papers are discussed and the results of both papers are laid down as an indication of the importance of volume electron microscopy in future research in this field.
Nanotomy of the Islets of Langerhans
Visualizing and analyzing the anatomy of pancreatic cells on the nanoscale is key to get a better view on how type 1 diabetes (T1D) progresses over time and to research the possibilities of treatment or cure. In 2013, Ravelli et al. performed large-scale electron microscopy (EM) to produce a larger field of view of the pancreatic rat cells [1]. They imaged the nano-anatomy (nanotomy) of the Islets of Langerhans to acquire accessible data that supports unbiased analysis.
Sample preparation is a crucial aspect of nanotomy, due to the often unique nature of the samples. With light microscopy the Islets, which only comprise around 1% of the whole pancreas, are localized to be sectioned on special carriers. After the quality of the samples is checked with EM, the samples are imaged with automated EM and the separate sections are stitched together. In this research, six separate datasets were recorded, each representing a different stage of T1D progression. In total the nanotomy data in this research comprises 25.000 images of pancreatic tissue, including information on cells, organelles and macromolecules. After analyzing the Islets of Langerhans during the progression of T1D in this rat model, the researchers detected glycogen accumulation in beta cells and other rare phenomena, for instance nuclear fibers resembling actin and cells undergoing mitosis.
Nanotomy creates substantial datasets where macrostructures of cells can be analyzed with high-resolution. Moreover, this information is accessible online, giving researchers all over the world the possibility to analyze and use this data. This technique could easily be used for human tissue and in other research fields, such as cancer research.
Network of Pancreatic Organ donors with Diabetes (nPOD)
In 2019, de Boer et al. saw the advantage to use the donors collected by the Pancreatic Organ donors with Diabetes (nPOD) program to get a clearer picture on the progression of diabetes in human pancreatic tissue [2]. The nPOD program collects from donors with T1D as well as non-diabetic donors with T1D-associated autoantibodies. Moreover, the organization also collects samples from transplantation-quality pancreatic donors. These samples are perfect to use as a control and therefore making this database the perfect source for a nanotomy project.
Within the nPOD program a standardized EM workflow was developed, circumventing several disadvantages of large-scale EM, and ensuring a high data quality for all samples. Overall the obtained database includes data from 47 donors, in which islets and neighboring exocrine tissue can be analyzed freely around the world. In total, the circa 1 million EM images contain larger structures, such as organelles like the Islets, but also smaller structures like macromolecules.
The results show that T1D donor samples contain numerous neutrophils, eosinophils and mast cells, both located in the islet and in the exocrine tissue. This might indicate that during the progression of type 1 diabetes not only the islets, but also the exocrine pancreatic tissue is affected. The role they might play is still unknown, but with these kinds of research new theories can be formulated on how diseases such as diabetes type 1 progress over time and what factors could play a role as possible targets for a cure.
Large-scale electron microscopy in the future
The technique of nanotomy is widely applicable and since the data is accessible online it has a high potential to become one of the main sources of information for nanoscale imaging. However, one of the main bottlenecks is the time it takes to collect and analyze each image. Researchers predict that this problem will become less pressing in the near future as developments are being made on automated image analysis. Automatically categorizing cell types based on their morphological appearance will definitely speed up the total EM workflow.
Furthermore, this database is still mainly focused on the acquisition and analysis of 2D samples. It would be highly recommended to acquire 3D information on this dataset to increase the understanding of underlying mechanisms of the T1D disease progression. High-throughput EM techniques, such as FAST-EM system, are being developed to improve research workflows and acquire more data in less time.
Would you like to know how FAST-EM will boost your workflow? Watch our recent launch demonstration to see what FAST-EM can do for you.
References
[1] Ravelli, R. B. G., Kalicharan, R. D., Avramut, M. C., Sjollema, K. A., Pronk, J. W., Dijk, F., Koster, A. J., Visser, J. T. J., Faas, F. G. A., & Giepmans, B. N. G. (2013). Destruction of Tissue, Cells and Organelles in Type 1 Diabetic Rats Presented at Macromolecular Resolution. Scientific Reports, 3(1). https://doi.org/10.1038/srep01804
[2] de Boer, P., Pirozzi, N. M., Wolters, A. H. G., Kuipers, J., Kusmartseva, I., Atkinson, M. A., Campbell-Thompson, M., & Giepmans, B. N. G. (2020). Large-scale electron microscopy database for human type 1 diabetes. Nature Communications, 11(1), 1. https://doi.org/10.1038/s41467-020-16287-5
.png)