The usage of electron microscopy (EM) in life science research has experienced significant growth over the past decade. While cryo-EM became the main technique for structural biology to reconstruct protein and macromolecular structures, novel volume-EM techniques are increasingly used to study large tissue samples and multicellular organisms [1][2].
These techniques are often not chosen for their ease of use, but rather for their ability to provide insights into unprecedented ultrastructural details of samples in three dimensions. This capability greatly enhances research in fields such as cell biology, developmental biology, pathology, and virology [2][3][4].
However, 3D EM techniques generate large and complex data sets, and the resulting images are often influenced by variations in parameter settings and workflows. It typically requires months to obtain validated results that can be put into scientific articles [5]. This is where data sharing and transparency become crucial.
The importance of data sharing
By enabling access and reuse of 3D EM datasets among the cryo-EM community, researchers can invite others to validate their reconstruction and compare them with known structures. This allows them to analyze conformational changes, observe the impact of mutations on the ultrastructure, and compare newly discovered viruses with known ones.
In the volume-EM community, researchers often explore connectomics or the ultrastructure of organelles. Access to raw 3D EM data allows them to compare their samples with archived samples to identify the differences and aid the development of segmentation models that require large datasets.
In educational settings, institutes may desire freely available datasets to train their staff or external students.
Valuable resources would include cryo-EM data for subtomogram averaging training, volume EM data for segmentation model building training, and CLEM data for correlation training. Moreover, stored data can serve also standard reference data for developing and testing new algorithms, including those related to data processing (e.g. stitching, 3D alignment), machine learning, deep learning, and correlative analysis.
Lastly, publicly available datasets can also drive the growth of knowledge and encourage collaboration. A research group may have acquired a volume-EM data set to address their specific research question, but there might be additional information within the dataset that can lead to novel insights. By leveraging the expertise and findings of others, researchers can make advancements in different research areas.
Public EM data archives
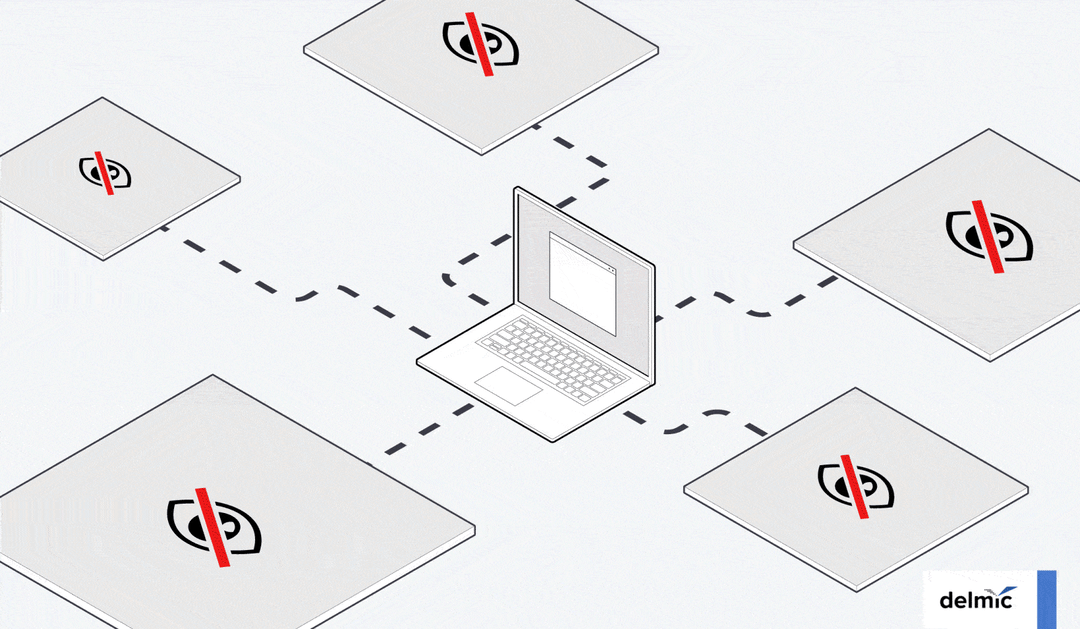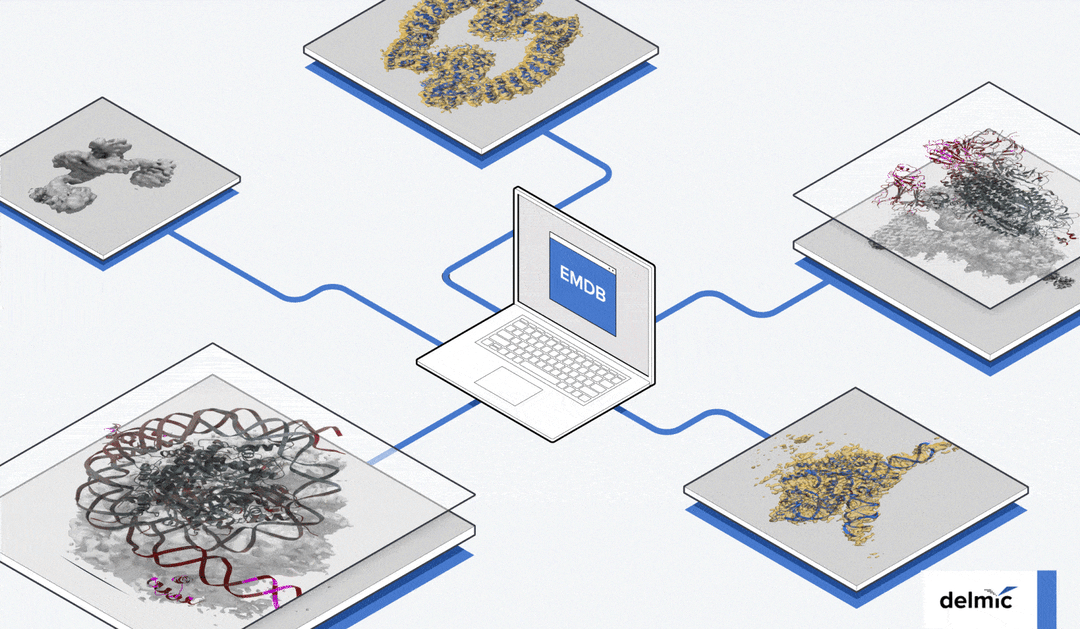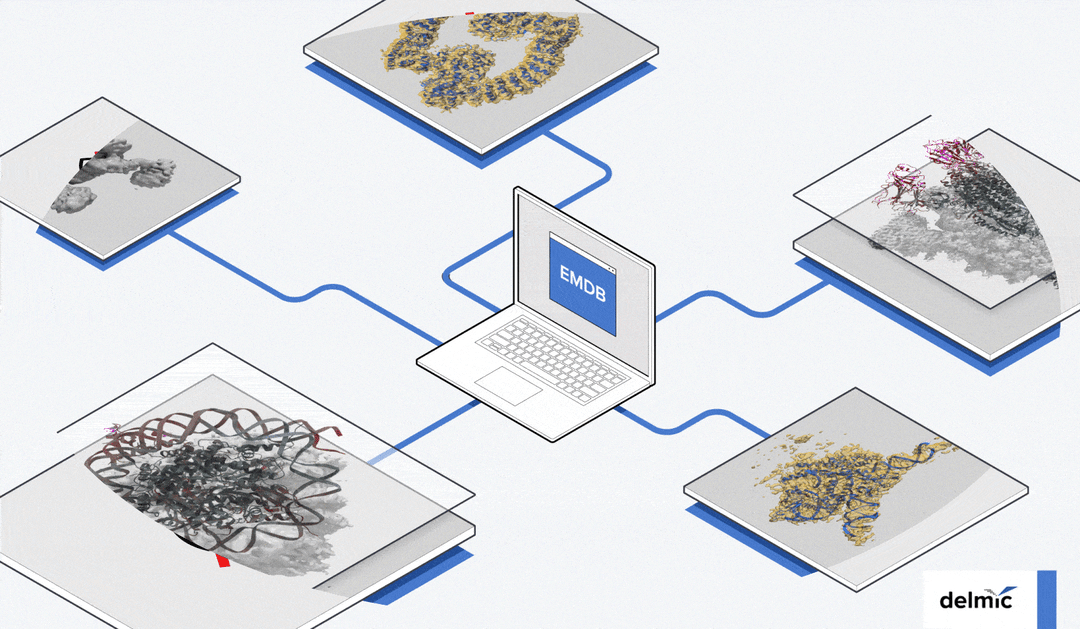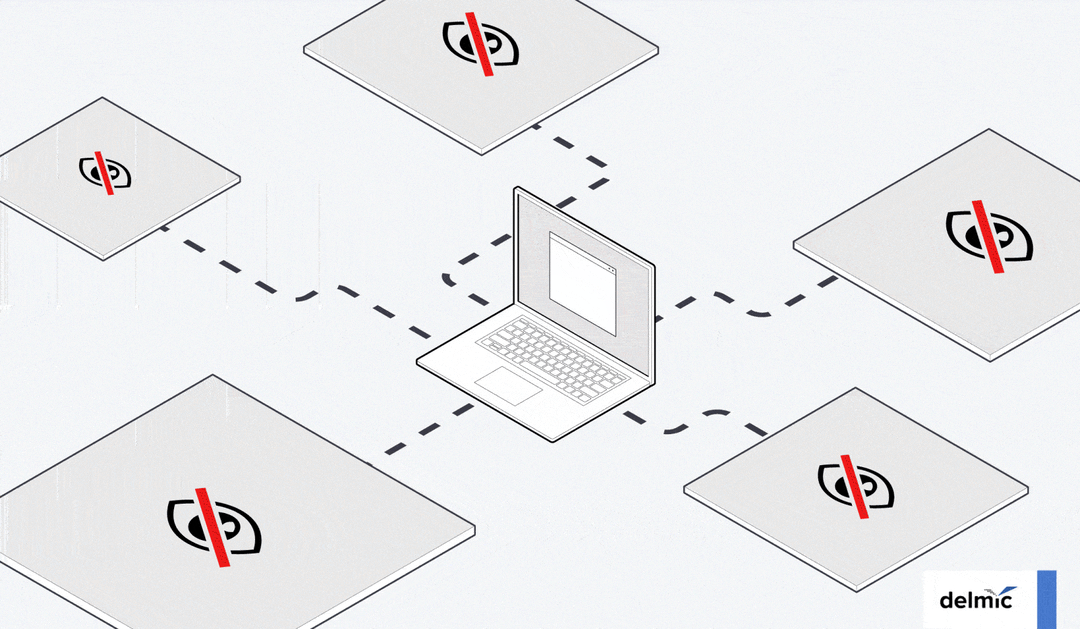
Despite these advantages, published research often doesn’t provide all the raw data, and copyright regulations generally restrict the reuse of published data. Fortunately, public image archives have been developed. Different EM data archives have been established for various purposes. The Electron Microscopy Databank (EMDB) archives 3D reconstructions from cryo-EM experiments, allowing users to easily compare their data with known 3D reconstructions [6][7] (Figure 1).
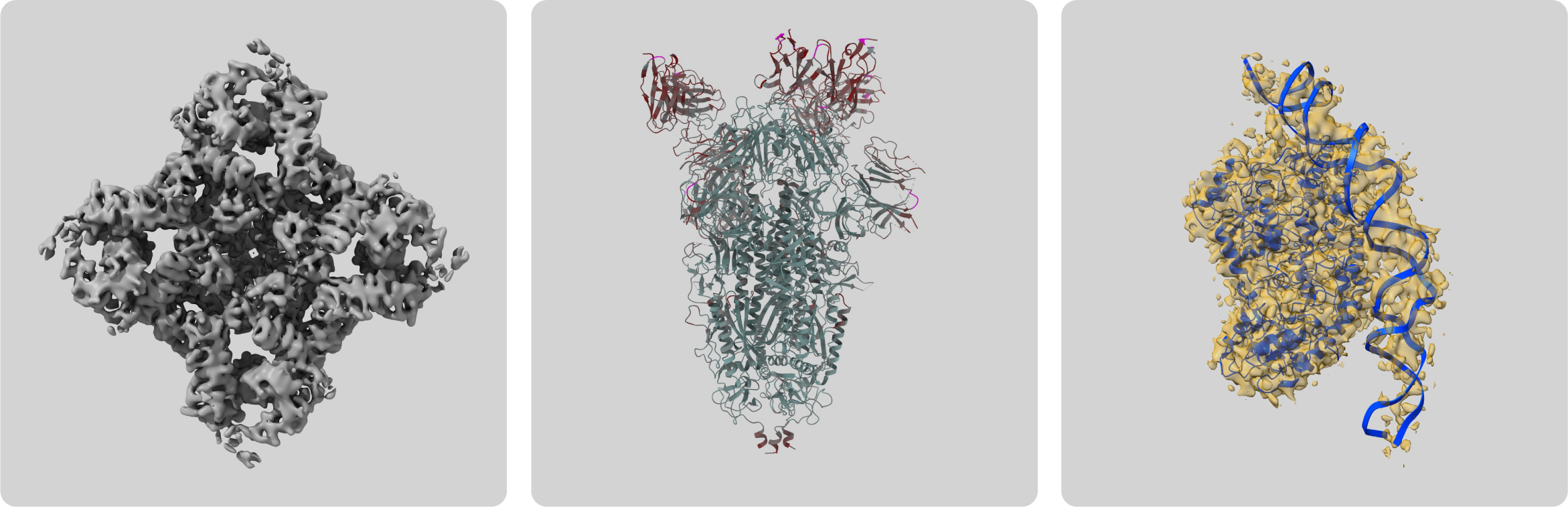 Figure 1: Examples of cryo-EM 3D reconstructions on the EMDB public repository [6].
Figure 1: Examples of cryo-EM 3D reconstructions on the EMDB public repository [6].
However, if the newly obtained reconstruction significantly differs from the archived reconstructions, it’s difficult to find out the reason for these differences. This is why The Electron Microscopy Public Image Archive (EMPIAR) was established in 2013 [5][8]. EMPIAR is a publicly accessible repository that provides raw 3D EM data, which underlies the data stored in EMDB. The archive includes data from a range of imaging modalities such as 3D cryo-EM, cryo-ET, volume-EM, and X-ray tomography [8].
The archived data is freely available for use without any restrictions, as it falls under the CC0 license.
This license allows the copyright holder to opt out of all copyright claims, rather than choosing from a range of permissions while retaining copyright [9]. Therefore, researchers can use the EMPIAR archive for various purposes, including method development, validation, experimental reuse, and training.
Micrographs or it didn’t happen
Since its establishment 10 years ago, the EMPIAR archive is widely used among researchers to share their research for validation and reuse. The data is also used as training data to enhance and teach data analysis tools and workflows [10].
Furthermore, on Twitter, where researchers often share and discuss findings from published scientific articles, the importance of sharing raw data is often emphasized. In one tweet, researchers asked the authors of a paper to make their raw data available, stating: ‘Micrographs or it didn’t happen #EMPIAR’.
Micrographs or it didn't happen #EMPIAR https://t.co/VMsFHvf1ju
— Paul V Thomas (@paul_v_thomas) February 19, 2020
Public image archives such as EMDB and EMPIAR have proved to be highly valuable resources in the field of electron microscopy. Furthermore, the accessibility of the archived data is in line with the movement of open-access scientific articles and open-access software. As the use of 3D EM continues to grow, data sharing and transparency will become of even greater importance. Therefore, public image archives for EM data will play an increasingly important role in driving scientific progress in the fields of life sciences and drug development.
References
[1] de Oliveira, T.M. et al., SLAS Discovery 26, 1, 17-31 (2021)
[2] Peddie, C.J. et al., Nat. Rev. Methods Primers 2, 51 (2022)
[3] Cyrklaff, M. et al., FEMS Microbiology Reviews 41, 6, 828–853 (2017)
[4] Benjin, X. et al., Protein Science 29, 872-882 (2020)
[5] Iudin, A. et al., Nucleic Acids Res. 51, D1503-D1511 (2023)
[6] The EMDB repository
[7] Lawson, C.L. et al., Nucleic Acids Res. 44, D396-D403 (2016)
[8] The EMPIAR repository
[9] The CC0 license
[10] Blog post ‘Getting a 3.5 Å GPCR structure in 1.5 h’ by CryoCloud
.png)