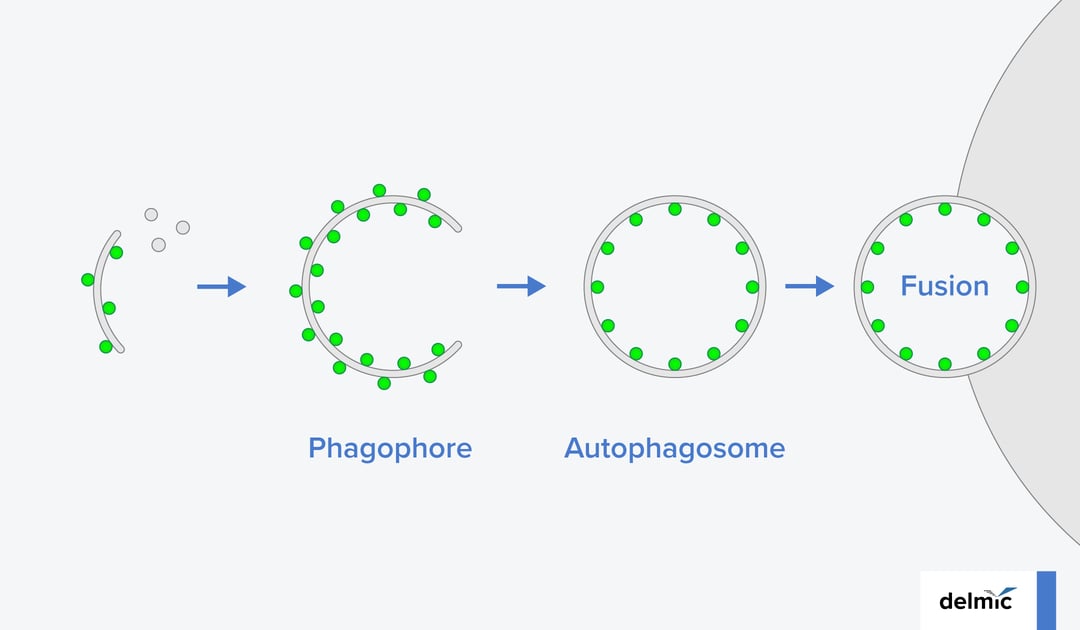
Autophagy is a process in which cells maintain their internal homeostasis, by clearing away debris such as dysfunctional proteins, damaged organelles, and intracellular pathogens. It starts with the formation of a double membrane vesicle, called an autophagosome. In the course of autophagosome development, the de novo synthesized membrane, called a phagophore, engulfs cellular material that needs to be discarded. Once the vesicle closes, it fuses with a lytic compartment where the waste material is digested.
The formation of autophagosomes has been researched with various biochemical and biophysical techniques. Up until recently the actual ultrastructural organization and dynamics of the membranes during autophagosome biogenesis were however not known.
To fill this knowledge gap, Bieber et al. applied in situ correlative cryo-electron tomography (cryo-ET) to monitor the autophagosome formation in its near-native cellular environment (1). By using this approach, the authors were able to visualize the membrane structures in autophagy within their near-native context at high resolution. Quantitative analysis of the 3D visualizations of these short-lived cellular structures revealed a number of new and unexpected insights into the biophysics of the autophagosome formation process.
The researchers were able to show and characterize distinct contact sites between autophagic structures and cellular organelles.
The high-resolution tomograms revealed that phagophores interacted differently with vacuoles than with the endoplasmic reticulum and the nuclear membrane. Although a clear physical connection with the latter two organelles was indicated in previous studies (2), Bieber et al. are the first to provide visual evidence and characterization of these contact sites.
Armed with the sub-nm resolution provided by cryo-ET, the researchers were moreover able to quantify several features of the autophagic membranes at various stages of autophagosome formation. As a result, the authors found that the intermembrane distance in these membranes is significantly smaller when compared to other structures in the cell, making it a unique trait that can aid in the visual identification of autophagosomes in the crowded intracellular environment.
A great challenge in this study was to visualize intermediate structures that are short-lived and therefore occur rarely in the samples. One of the strategies to overcome this problem was the application of cryo-correlative light and electron microscopy (cryo-CLEM). To make use of this method, the authors fluorescently labeled the specific structures of interest and then detected them using a cryo-fluorescence light microscope (cryo-FLM). This approach played an important role at two stages of the cryo-ET workflow: sample milling and tomogram acquisition.
Sample milling takes place typically in a cryogenic focused ion beam/scanning electron microscope (FIB/SEM) setup.
Using the FIB, a sample is thinned down to 100-300 nm so that during transmission electron microscopy (TEM), electrons can pass through the sample and form an image. Without guidance as to where the regions of interest (ROIs) are, they can be easily milled away, rendering the lamellae useless and making the cryo-ET workflow extremely time and labor-intensive. By using METEOR, a fluorescent light microscope directly integrated into the cryo-FIB/SEM, the researchers were able to check the presence of the ROIs during and after the milling process (3). After obtaining the lamellae, the researchers overlaid and correlated the cryo-fluorescence images with the TEM maps.
In this way, they were able to identify autophagic structures and choose the correct ROI for tomogram acquisition.
In summary, the work by Bieber et al. is a great example of how cryo-CLEM can be successfully applied to study dynamic, short-lived ultrastructures in the native and complex cellular environment. At Delmic, we are excited to see how this approach will further advance our understanding of cellular organelles formation.
References
- Bieber A., et al. (2022) In situ structural analysis reveals membrane shape transitions during autophagosome formation. PNAS, 10.1073/pnas.2209823119
- Graefa, M., et al (2013) ER exit sites are physical and functional core autophagosome biogenesis components Mol. Biol. Cell doi.org/10.1091/mbc.e13-07-0381
- Smeets, M., et al. (2021) Integrated Cryo-Correlative Microscopy for Targeted Structural Investigation In Situ. Microscopy Today, doi:10.1017/S1551929521001280
.png)