For this reason, overlaying images from different microscopes is often a chosen method for researchers, particularly in the life sciences. What is frequently combined is data from an electron microscope (EM) and a fluorescence microscope (FM). These images are then often “overlayed” for a complete picture of the data.
This is, however, a great challenge. The user needs to deal with the massive difference in field of view and resolution between the optical and electron microscopes, as well as distortions on the sample that can occur as a result of exposure to an electron beam [1]. For this reason, several methods exist to achieve overlay, some more effective than others. More recent technological developments in microscopy are able to achieve extremely accurate overlay.
In FM, samples are prepared to emit fluorescent light, either by staining, immunofluorescence, or with the use of fluorescent proteins. EM, on the other hand, provides high-resolution images at the nanoscale for contrast, which opens up essential information on the structure of a cell, organelle, or organic tissue. Combined, these results thus offer high-resolution black-and-white images with colors that serve as markings for particular parts of interest within the sample.
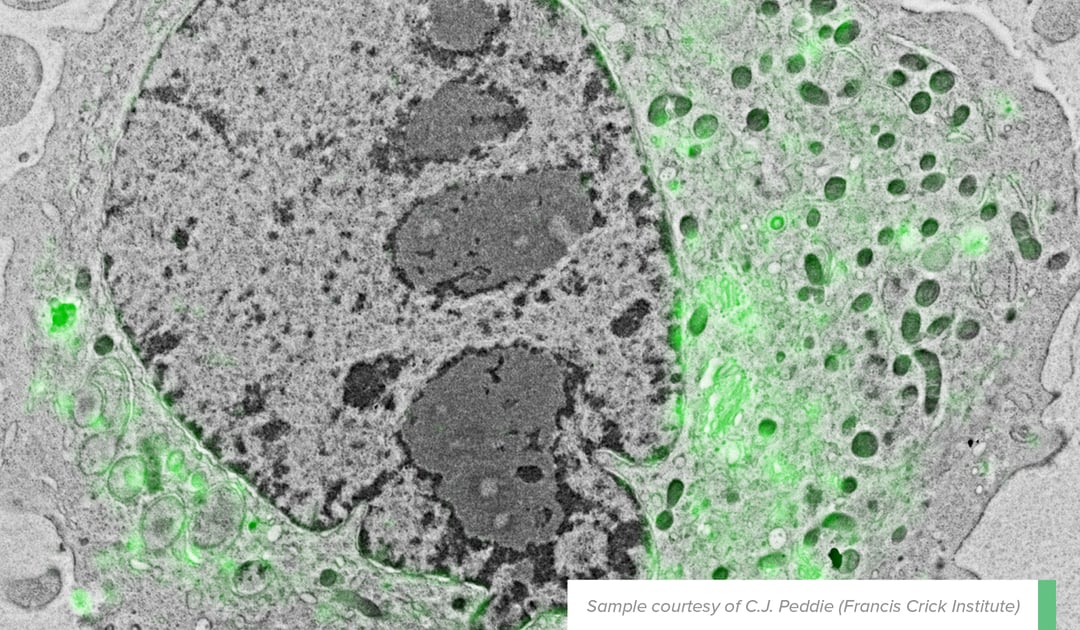
In an example that has been touched upon in a previous blog post, overlayed FM and EM images were able to show that the diacylglycerol (DAG), a protein involved in membrane morphology and dynamics in the nuclear membrane, is positioned at the “nuclear envelope, nucleoplasmic reticulum and curved tips of the Golgi apparatus” [2]. The EM image provided detailed contextual information needed to identify exactly where the protein – marked with GFP-C1 fluorescence signals – was located in the cell (see figure beside).
Figure: Localization of the lipid diacylglycerol within cellular membranes of HeLa cell expressing GFP-C1. Sample courtesy C.J. Peddie Francis Crick Institute
This example is an excellent illustration of the importance of overlaying images from two different microscopes to obtain detailed and accurate information regarding the location of biomolecules in a sample. The overlayed image was achieved with integrated correlative light and electron microscopy (integrated CLEM), a method widely considered to be the most effective and accurate tool for performing the overlay of microscopy data.
Integrated CLEM is the newest of many overlaying methods preceding it. With “shuttle and find”, for example, the researcher makes use of a sample holder that can be used in both FM and EM. This sample holder enables the use of markers to indicate the location of the region of interest on the sample. This method is not very exact, and has a precision of up to 10 micrometers at best. It is arguably not a method of overlay, but rather simply a means of finding the sample to begin with.
Another method is manually overlaying the images with the use of image editing software such as Photoshop. In this case, the researcher makes use of the layers and blending functionalities to combine the two images into one. Expertise in the research topic is used to identify where the images match up. Although this is a method that rarely goes wrong, it is nevertheless questionable. The accuracy of the overlay needs to be substantiated with other data, which is only effective to a certain degree. In this case, the bias of the scientist always risks coming into play.
One way to make this method more accurate is to use multiple color channels. This provides more information and distinguishes different objects from one another. However, like shuttle and find, this is more useful as a means of identifying the region of interest rather than to collect detailed and accurate data.
The risk of bias is greatly reduced when fiducial markers are used. In an exemplary case in point, Kukulski et al. [1] placed several “fluorescent microspheres” on a section prior to imaging. These serve as markers that can be visualized by both the electron and fluorescent microscopes, and are able to account for changes in resolution and distortions between the two microscopes. This is widely considered a very robust means of correlating visual data. It is however, problematic in terms of being extremely complicated and time-consuming. A high level of specialization is required to carry out the placement of the tiny fiducial markers by the researcher. Bias can also come into play, as the researcher risks confusing parts of a cell with a marker.
Ultimately, the most effective method in terms of accuracy and ease of use is an integrated system that combines FM and EM into one, or CLEM. This is the method used by Peddie et al. for their research on the nuclear membrane. In such a system, the two microscopes take images simultaneously and overlay is carried out automatically. Identifying the region of interest is no longer a part of the methodology to begin with, as the system ensures that the user is looking at the same region at the same time with both microscopes.
This method of automated overlay uses a grid of temporary, non-invasive fiducial markers to align the images and compensate for changes in rotation and resolution. This is achieved using cathodoluminescence, in which the light generated by electrons when they hit a luminescent material can be detected using the fluorescence microscope. This is a fully mechanical procedure that eliminates the risk of bias.
It is of essence for the scientist to distance him or herself as much as possible from the object of research with the use of the right methods and technology. In the case of overlaying visual data in the life sciences, one must choose a method that can achieve the greatest possible accuracy in the fewest possible steps. The use of more accurate tools not only adds to the integrity of the research, but also leaves the necessary time and energy for new scientific inquiries.
References
[1] Kukulski, Wanda, et al. "Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision." The Journal of cell biology 192.1 (2011): 111-119.
[2] Peddie, Christopher J., et al. "Correlative and integrated light and electron microscopy of in-resin GFP fluorescence, used to localise diacylglycerol in mammalian cells." Ultramicroscopy 143 (2014): 3-14.
If you would like to learn more, you can download our technical note on automated overlay:

Automated overlay
.png)