In particular, you may be interested in examining biomolecules and their function within the greater context of the cell as a whole. Recent technological and methodological developments in microscopy have made this process much more straightforward, with integrated fluorescence and electron microscopy. Such a system allows for automatically overlayed images from both an electron and a light microscope, providing you with the ability to identify certain organelles or biomolecules by tagging, at the same time that you are able to localize where they are situated within the cell. More recently, super-resolution fluorescence imaging has been developed, which opens up even greater opportunities for learning about the complexities of life.

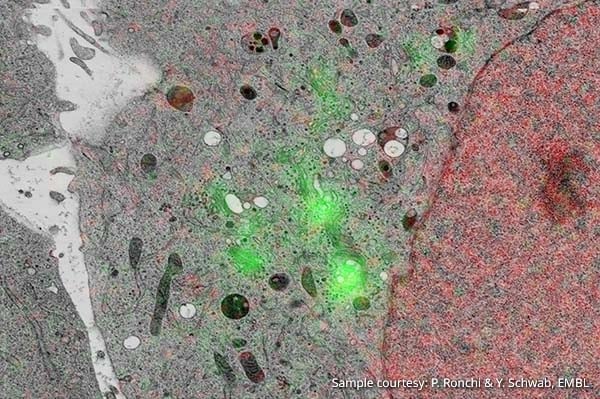
Image: Hela Kyoto cells stably expressing GalNAC-T2-GFP and Histone 2B-mcherry. The cells were grown on carbon coated sapphire disks and high pressure frozen. The cells were then freeze substituted with 0.1% UA in glass distilled acetone in Lowicryl HM20.
Sample courtesy: P. Ronchi & Y. Schwab, EMBL
One of the primary tools for studying cells at small length scales remains fluorescence microscopy, due to its speed and efficiency. With this method, perhaps familiar to all life scientists reading this, molecule-specific labelling is used to mark the cellular components of interest under fluorescent light, with a limited impact to the living sample.
The main disadvantage to fluorescence microscopy, however (and other forms of light microscopy, for that matter), is the diffraction barrier. To put it in simple terms, this imposes a limit upon the ability to distinguish between two different objects that are approximately less than half the wavelength of the light used (thus, in the case of FM, less than half the wavelength of fluorescent light) apart from one another. Ultimately, this means that objects smaller than 200nm cannot be visualized with typical fluorescence microscopy approaches.
As you probably know, crucial to research in the life sciences is a study of proteins and their function in the cell. However, proteins are typically around 4 nanometers in diameter; too small for a conventional fluorescence microscope (Watanabe et al., 2011). In order to overcome this problem, super-resolution (SR) fluorescence microscopy has been developed as an alternative. This method uses innovative technology to transcend the diffraction limit and thus to image samples at a resolution of up to 20nm, which was previously only attainable with an electron microscope.
Ever since its invention, SR microscopy has been widely used in the life sciences. In particular, this method is used to more precisely locate and to gain a deeper understanding of specific biomolecules. Nevertheless, even what can be visualized through SR microscopy is limited. Ultimately, the researcher is left with a black backdrop for the area that has not been labelled, thus providing no information regarding the cell’s ultrastructure.
For research that requires in-depth knowledge of cellular processes, electron microscopy (EM) is thus used to understand the micro-environment in which biomolecules operate. Ideally, EM is used in combination with fluorescence microscopy, a solution otherwise known as correlative light and electron microscopy (CLEM). Correlative microscopy allows for the precise localization of fluorescent labels with the fluorescence microscope, while the electron microscope delivers detailed structural information of the surrounding environment. The combination of EM with super-resolution fluorescence microscopy (sometimes referred to as super CLEM) is even more effective for closely examining biomolecules within their contexts with its extremely high resolution and accuracy.
One example of an application of CLEM in the life sciences is a study which identified GFP-tagged proteins in combination with electron microscopy. It was found that diacylglycerol (DAG), a protein involved in membrane morphology and dynamics in the nuclear membrane, is positioned at the “nuclear envelope, nucleoplasmic reticulum and curved tips of the Golgi apparatus” (Peddie et al., 2014, pg. 3). This finding demonstrated how effective this technology is for identifying specific biomolecules in a cell and thus for revealing crucial information regarding their function. While this research attested to the value of a system that combines fluourescence microscopy with electron microscopy, the authors nevertheless state that they struggled with the limitations imposed by the diffraction barrier. They would thus potentially have benefited more from a super-resolution microscope.
In another study using super-resolution fluorescence microscopy in combination with electron microscopy, researchers successfully visualized the location of proteins of nuclear pore complexes in the oocyte ofXenopus laevis, or an African clawed frog. With the use of super-resolution CLEM, they were able to “map the position of proteins in the context of the ultrastructure of large multiprotein complexes” (Löschberger,et al, 2014, pg. 2). As a final case in point, yet another study was able to successfully employ super-resolution correlative microscopy by identifying a previously unknown position of the protein epsin, which is essential to the formation of vesicles on the plasma membrane (Sochacki et al., 2014).
Evidently, correlative microscopy, and that using super-resolution fluorescence imaging more specifically, is an extremely important tool, essential for gaining a deeper understanding of the vast complexities of life. On that note, we leave you with a list of references for which super-resolution correlative microscopy was used. We also invite you to download the brochure for our latest release, the SECOM SR, a high-performance correlative light and electron microscope with super-resolution fluorescence imaging technology.
Further reading
Chang, Yi-Wei, et al. "Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography." Nature methods 11.7 (2014): 737-739.
Johnson, Errin, et al. "Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins." Scientific reports 5 (2015).
Kopek, Benjamin G., et al. "Correlative photoactivated localization and scanning electron microscopy."PLoS One 8.10 (2013): e77209.
Ligeon, Laure-Anne, et al. "Structured illumination microscopy and correlative microscopy to study autophagy." Methods 75 (2015): 61-68.
Liss, Viktoria, et al. "Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy." Scientific reports 5 (2015).
Löschberger, Anna, et al. "Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution." J Cell Sci 127.20 (2014): 4351-4355.
Paez-Segala, Maria G., et al. "Fixation-resistant photoactivatable fluorescent proteins for CLEM."Nature methods 12.3 (2015): 215-218.
Perkovic, Mario, et al. "Correlative light-and electron microscopy with chemical tags." Journal of structural biology 186.2 (2014): 205-213.
Sochacki, Kem A., et al. "Correlative super-resolution fluorescence and metal-replica transmission electron microscopy." Nature methods 11.3 (2014): 305-308.
Watanabe, Shigeki, et al. "Protein localization in electron micrographs using fluorescence nanoscopy."Nature methods 8.1 (2011): 80-84.
References
Löschberger, Anna, et al. "Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution." J Cell Sci 127.20 (2014): 4351-4355.
Peddie, Christopher J., et al. "Correlative and integrated light and electron microscopy of in-resin GFP fluorescence, used to localise diacylglycerol in mammalian cells." Ultramicroscopy 143 (2014): 3-14.
Sochacki, Kem A., et al. "Correlative super-resolution fluorescence and metal-replica transmission electron microscopy." Nature methods 11.3 (2014): 305-308.
Watanabe, Shigeki, et al. "Protein localization in electron micrographs using fluorescence nanoscopy."Nature methods 8.1 (2011): 80-84.
.png)