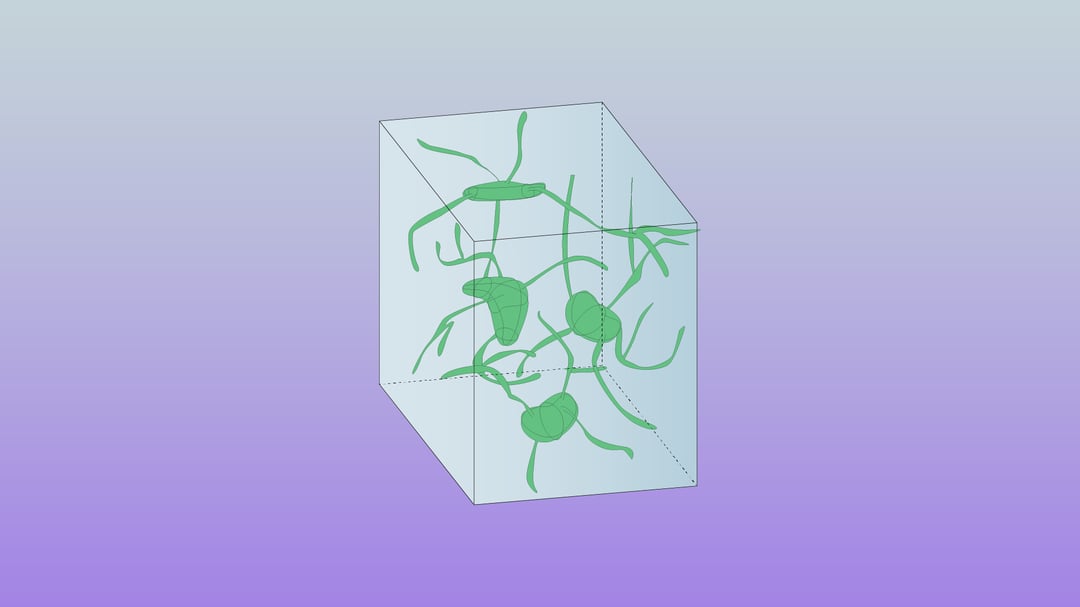
Recent years have seen a rapid increase in the field of connectomics, the study of tracing and understanding connectivity maps of the brain. Thanks to its ability to resolve the level of detail required to create these maps, electron microscopy has become an essential tool for the mapping efforts undertaken in connectomics [1].
The capabilities of electron microscopy in connectomics have evolved drastically these past few years. Technological advancements in automation and robustness have made projects possible that aim to image and map brain structures and even whole small brains [2,3]. Despite this progress, creating these fascinating datasets (connectomes) remains a daunting task due to several limiting factors during the process of sample preparation, imaging and data analysis.
Sample Preparation
The collection of data requires several steps, which need to be completed carefully and thoroughly to get a detailed 3D image of the brain structures. The workflow starts with sample preparation. Sample preparation is time-consuming because the ultrastructure of the tissue needs to be preserved and uniformly stained. Samples are then sliced into thin layers known as sections, for which an (automated) ultramicrotome is used. Techniques have been developed to make this step less laborious and more efficient [4,5].
Imaging
Electron imaging of all the sections is probably the most limiting factor in connectomics, as the throughput of an EM defines the size of the project. As mentioned in the blog post about volume electron microscopy, the time needed to record massive datasets in volume EM can go up to weeks, months or even years and this is not different for large-scale projects such as connectomics.
Data Analysis
After the image acquisition, a 3D representation of the sample is made. This 3D volume serves as the basis for connectivity research, as it contains the data required to map out the neural connectivity. Due to the scale of the data, analysis and annotation are rather time-consuming, and require substantial manpower to complete. However, as discussed in the previous FAST-EM blog post, advances in computing power and the contributions of large research groups lead to new possibilities, where even larger volumes of samples can be analyzed within a reasonable time frame.
FAST-EM
The FAST-EM system developed at Delmic, together with Thermo Fisher, Technolution and TU Delft, reduces the time-consuming aspect of imaging by dealing with the issue from different angles. Delmic’s FAST-EM uses 64 electron beams, allowing faster, parallel imaging of a sample and therefore increasing the throughput. Another advantage is the use of STEM (Scanning Electron Transmission Microscopy), which leads to a decrease in dwell time per pixel (as short as 400ns) and still yields an excellent signal-to-noise ratio for each image. Two other features of the FAST-EM system that increase the throughput are the sample carrier and the automation of the system. The carrier can hold up to 9 substrates, each of which can contain hundreds of sections, and the system uses software that is robustly automated and easy to use. These features enable an independent workflow with no need of supervision lasting for up to 72 hours.
All these improvements in the electron microscopy workflow, give rise to new opportunities for connectomics, but also other research projects. The decrease in time to acquire large 3D images enables imaging of multiple specimens simultaneously. This opens up the possibility to compare the same specimen under different experimental conditions, like genetic modification, or drug or ablation treatments.
References
[1] Briggman, K. L., & Bock, D. D. (2012). Volume electron microscopy for neuronal circuit reconstruction. Current Opinion in Neurobiology, 22(1), 154–161. https://doi.org/10.1016/j.conb.2011.10.022
[2] Shan Xu et al (2020). A Connectome of the Adult Drosophila Central Brain. bioRxiv 2020.01.21.911859. https://doi.org/10.1101/2020.01.21.911859
[3] Zheng, Z., et al (2018). A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell, 174(3), 730-743.e22. https://doi.org/10.1016/j.cell.2018.06.019
[4] Titze, B., & Genoud, C. (2016). Volume scanning electron microscopy for imaging biological ultrastructure. Biology of the Cell, 108(11), 307–323. https://doi.org/10.1111/boc.201600024
[5] Mikula, S., Denk, W. High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nat Methods 12, 541–546 (2015). https://doi.org/10.1038/nmeth.3361
.png)