Two interesting research papers that described the replication environment of SARS-CoV-2 will be discussed in this post. Both papers used cryo-ET and subtomogram averaging to image important molecules and structures that play a crucial role in the replication of the coronavirus genome.
Researching induced DMV molecular pore complexes
The first study we would like to cover here was conducted by Wolff et al. in which they used murine hepatitis coronavirus as a model for SARS-CoV-2 to better understand the coronavirus replication mechanism [1]. Their paper revealed the presence of pore complexes on the membrane of virus-induced cytosolic double-membrane vesicles (DMVs) [1]. These complexes were proposed to act as a passageway through which the viral mRNA and synthesized viral genome can travel between the interior of the DMVs and the cytosol of the host cell. In the cytosol, the mRNA and viral genome can be translated and assembled into progeny virions.
After preparation with focused ion beam milling, cryo-lamellae were imaged with cryo-ET. The tomographic slices indeed showed pore complexes inside the membranes of the DMVs. Sixfold-symmetrized subtomogram averaging gave more insight and, after 3D surface rendering, assisted in unraveling the 3D architecture of the pores embedded in the membrane of the DMVs. Even though the purpose of the pores still remains to be elucidated, they could provide a coronavirus-specific drug target, as they most likely play a crucial role in the replication, and thus progression of this viral infection.
Imaging structure and replication of SARS-CoV-2
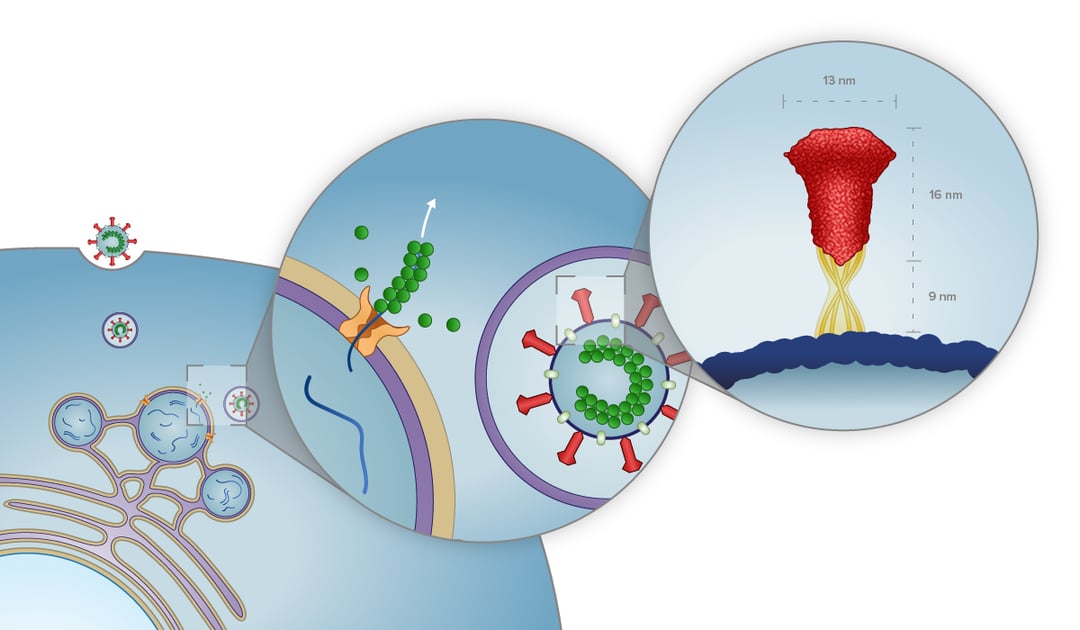
The second study we would like to discuss, which was conducted by Klein et al. using a SARS-CoV-2 strain, focused not only on in situ imaging of the viral replication compartments (or DMVs) but also the viral budding mechanism and the structure of extracellular virions [2].
Here, it was shown, through cryo-ET imaging that the DMVs indeed contain RNA molecules. However, it is still uncertain if the DMVs function as a replication site or as a way to separate ready-to-use RNA from the host cell environment. Moreover, the DMVs were shown to interact with each other and link membranes. Klein et al. suspect that this is part of the virion budding stage of the replication cycle of the virus.
Furthermore, by imaging the virion assembly with in situ cryo-ET it was possible to study the process in near-native conditions and localize individual S trimers and viral ribonucleoprotein complexes (vRNPs). The data collected indicate that the S trimers are remarkably motile and flexible, and can have angles of up to 41° with regard to the membrane. The vRNPs were also found to be associated with the membrane of virions and possibly enable efficient packaging of the relatively large genome of SARS-CoV-2.
Importance of cryo-ET in virology
As described in this blog post, cryo-ET is a perfect tool to image and study the architecture and environment of molecular complexes in a near-native state. In virology, these detailed images can aid researchers in their search for medicine targets or vaccines for viruses like COVID-19.
Check out METEOR, one of our cryo solutions microscopes, to get more information on the system and how it can boost your workflow!
References
[1] Wolff, G., Limpens, R. W. A. L., Zevenhoven-Dobbe, J. C., Laugks, U., Zheng, S., de Jong, A. W. M., Koning, R. I., Agard, D. A., Grünewald, K., Koster, A. J., Snijder, E. J., & Bárcena, M. (2020). A molecular pore spans the double membrane of the coronavirus replication organelle. Science, 369(6509), 1395–1398. https://doi.org/10.1126/science.abd3629
[2] Klein, S., Cortese, M., Winter, S. L., Wachsmuth-Melm, M., Neufeldt, C. J., Cerikan, B., Stanifer, M. L., Boulant, S., Bartenschlager, R., & Chlanda, P. (2020). SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nature Communications, 11(1). https://doi.org/10.1038/s41467-020-19619-7
.png)